|
||||||||||||||||||
|
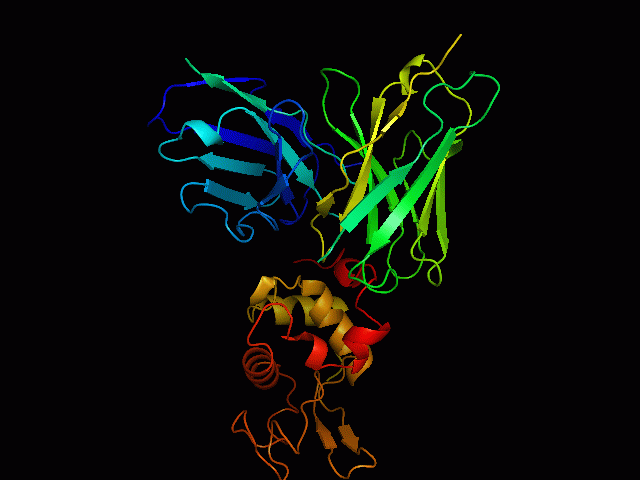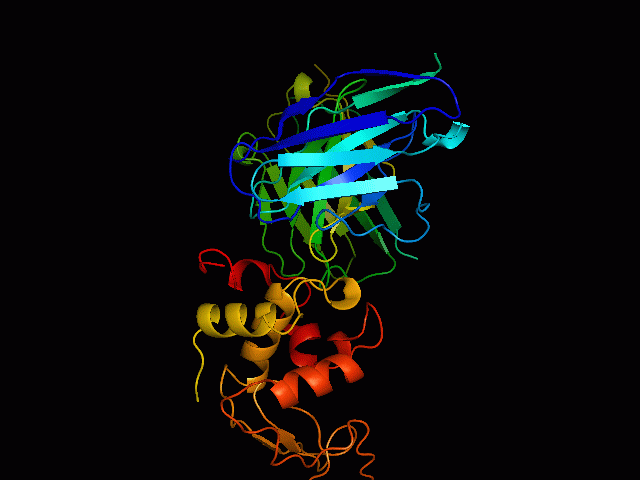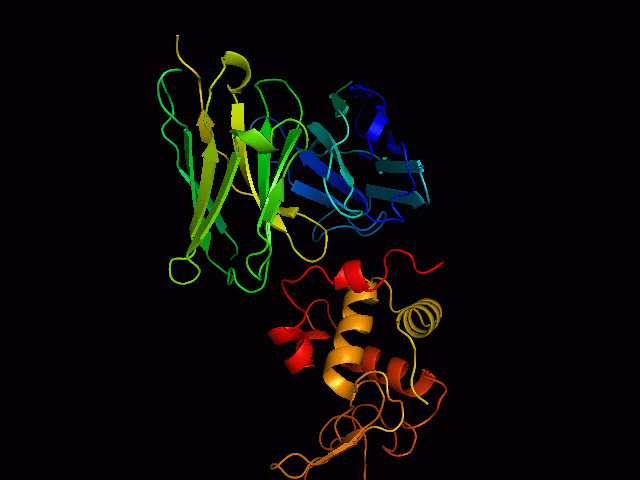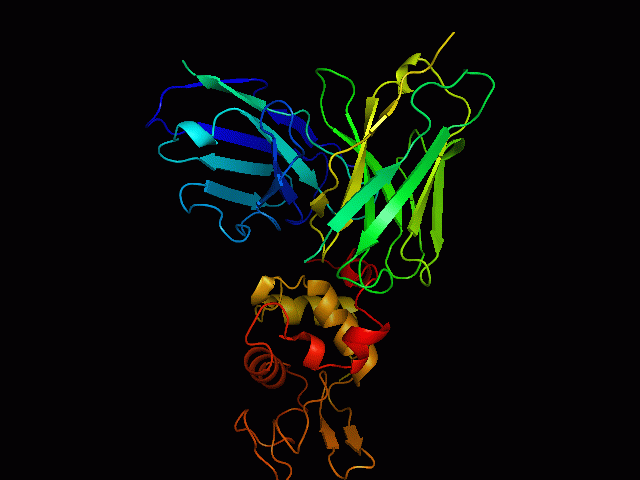
Discontinuous B Cell Epitopes Identification of conformational B-cell epitopes in antigens is an important key step in rational vaccine design. Computational prediction of the epitopes is a challenging problem in immunoinformatics. Most of the B-cell epitopes are known to bind to antibodies with sequentially discontinuous but conformationally continuous surface residues, hence structure based prediction methods are more efficient than sequence based methods. However, performance of existing prediction tools are not very encouraging, mainly due to limited number of available unique epitope structures and sequences. Use Of Support Vector Machine We have developed an epitope prediction method based on training Support Vector Machines (SVM) to develop prediction classifiers. The SVM models were trained with integrated features of sequence based physicochemical scales as well as structure based features of known epitopes. This method takes into account the surface accessibility, physicochemical properties of amino acids, and the secondary structures of the epitopes (helices, strands and loops). The training dataset used in the study consists of antigens extracted from 157 PDB structures of antigen-antibody complexes. Accuracies of the models was found to be 92%for RBF kernel. |
|
||||||||||||||||
|
|
|
|||||||||||||||||