

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| BBOV_I001480 | OTHER | 0.999536 | 0.000188 | 0.000275 |
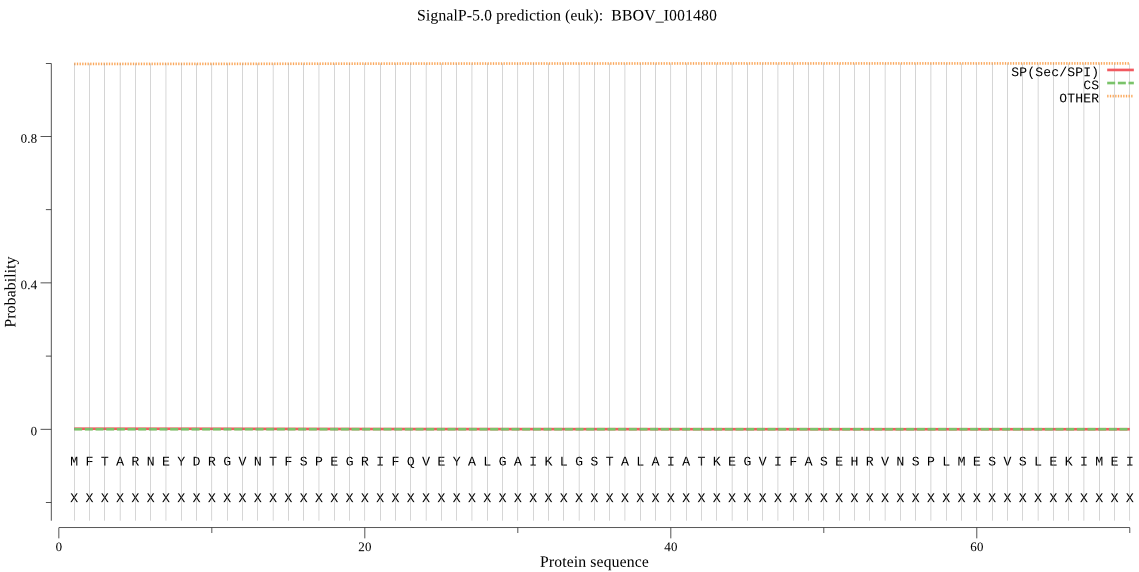
MFTARNEYDRGVNTFSPEGRIFQVEYALGAIKLGSTALAIATKEGVIFASEHRVNSPLME SVSLEKIMEIDTHIGCTMSGLIADARTLIDHGRLECANHRFVYNEPLAIRSCVESIADMA LDFSDIFDTRKKKTMSRPFGVALLVGGIDIDGPSIWCVDPSGTSIKYKASAIGSAQEGAE SVLQERYRDDMSFRDAELLVLEVLRQVMKDQMTTKNIEMARIKIHDRQFREYTEDEIGEI IQDLPQMENM
| ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|
| BBOV_I001480 | 192 S | RDDMSFRDA | 0.997 | unsp |