

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| BBOV_II000170 | SP | 0.085171 | 0.914568 | 0.000261 | CS pos: 32-33. VTA-DL. Pr: 0.6673 |
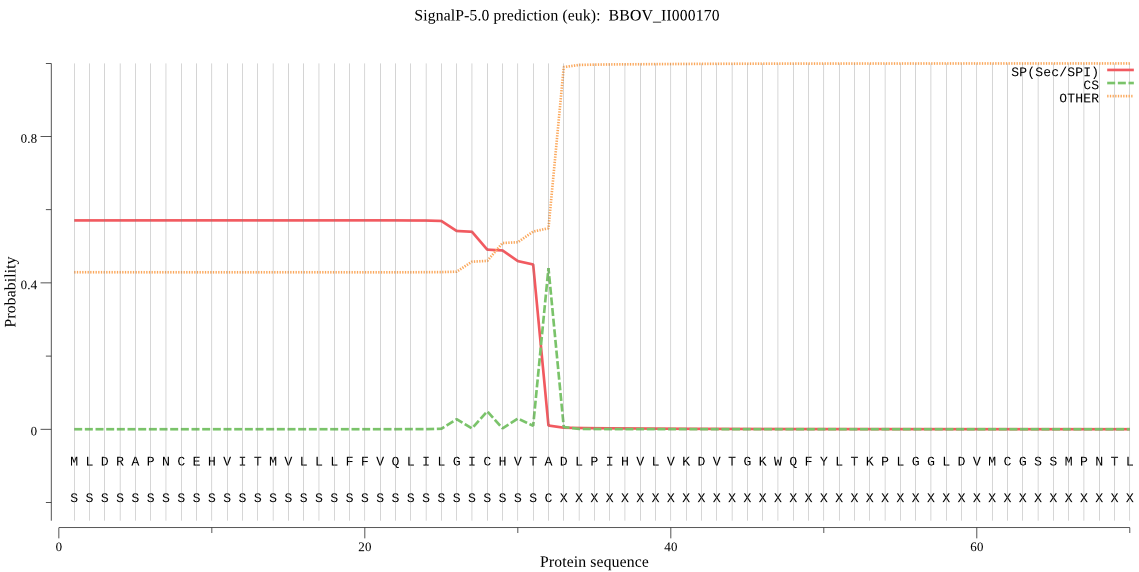
MLDRAPNCEHVITMVLLLFFVQLILGICHVTADLPIHVLVKDVTGKWQFYLTKPLGGLDV MCGSSMPNTLEGNLKLGNYLDYLKSHYTLDRNFQLELSLDVIPYSDTSNKPNRYKWRALA VKDAAGKVVGRWTIVNDQGFEVLLDDSTRYFFYIRYTQNGDMFETDPNHTQIGWAYRPGN LSAAVTERRCAFASKVNALTPPQTTLVKLHTNVTEMFKQIDRSITNNGTSHGAIKSAHIS PKRGLYPCECANKERLHFDDQLPVHFEWKTRATIPIVNQDSCGSCHAIASRYVLQSRFLI ALERMSNRSPELEGILDELSHYTFDPKDVTDCSMYNQGCDGGYPYLMGKQMREFGILTTK NAGQQCTLLSTERRYFARDYGYVGGCHQCTACQGDALIMREILANGPVVTAIDAAVLTAD YDGHIITSAEEGTNSGICDMEHHPILTGWEYTSHAVAIVGWGQEKVGARMIKYWICRNSW GQNWGINGHFKIERGKNAYGIESEAVFIDPDFSKFAQEPAASFLHTIHHH
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| BBOV_II000170 | 306 S | LERMSNRSP | 0.995 | unsp | BBOV_II000170 | 306 S | LERMSNRSP | 0.995 | unsp | BBOV_II000170 | 306 S | LERMSNRSP | 0.995 | unsp | BBOV_II000170 | 309 S | MSNRSPELE | 0.993 | unsp | BBOV_II000170 | 108 S | YSDTSNKPN | 0.992 | unsp | BBOV_II000170 | 240 S | SAHISPKRG | 0.996 | unsp |