

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
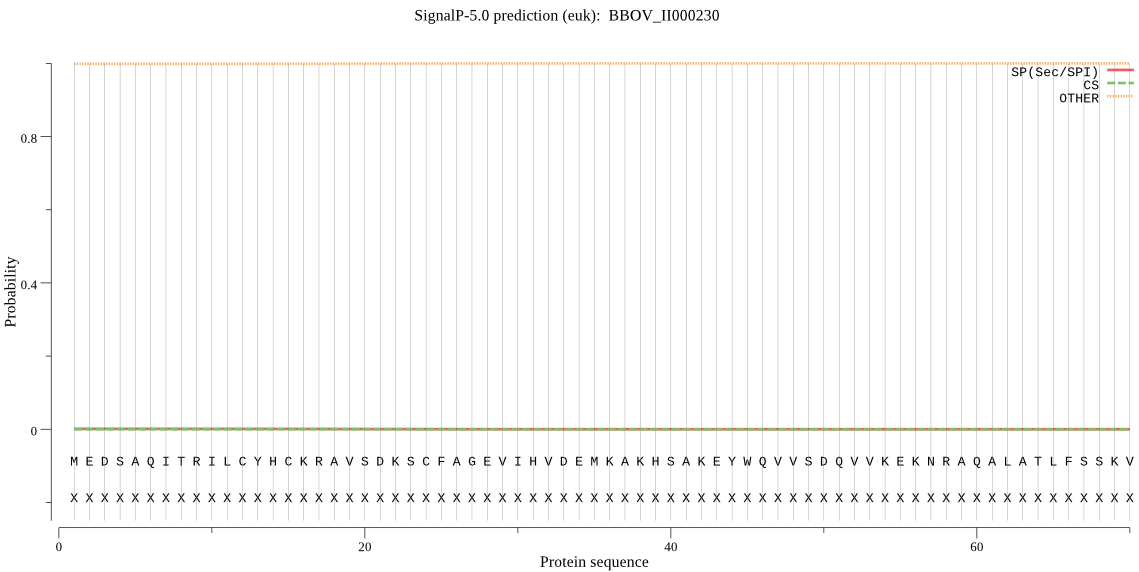
| BBOV_II000230 | OTHER | 0.999560 | 0.000365 | 0.000075 |
MEDSAQITRILCYHCKRAVSDKSCFAGEVIHVDEMKAKHSAKEYWQVVSDQVVKEKNRAQ ALATLFSSKVVEIFKFLGKTLVDIPINMENSKEMQFDLFSMPPSIRMCQECLCFFSIERS CHCESCMGPDHISHYVVLMHGVVADPLCMAFIAQSLLEVYPHLFIYFPHKIAGKSLVGLE LVVKTIGTELLELFSKIPRKIKLSIIGHSFGGVILRHWYFFYSRKTPGIHHYPKYSSASG STGEDSTIKQDGAEDENPREVPEIIWCNYMSVASPHAGIYENNAAFRKIVGLVGSKTVDE LDNDSVDLLFLASRESMDGMKKFKNVVVYGNLSGDFLVAPRTSLLMPRHRVAGKMVKSFI KYAEKDTGEPHSIWDMLSKCDFTQVGSDDESDSDVESSGSFDKDCIMAKLMESFYATVAS LIKKTQHLAMFTGKPYEPVNGESNEDLCKRYFTFDDLEFFKMLFSQIEDKSSKKLLSRLA ASPKLLYNEVLLTALWDMAPNKYAVYLPTLSLPHRSILTPLEGSLWHKYTNKVLPQIASI FIT
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| BBOV_II000230 | 241 S | SASGSTGED | 0.992 | unsp | BBOV_II000230 | 241 S | SASGSTGED | 0.992 | unsp | BBOV_II000230 | 241 S | SASGSTGED | 0.992 | unsp | BBOV_II000230 | 391 S | SDDESDSDV | 0.996 | unsp | BBOV_II000230 | 393 S | DESDSDVES | 0.995 | unsp | BBOV_II000230 | 443 S | VNGESNEDL | 0.991 | unsp | BBOV_II000230 | 20 S | KRAVSDKSC | 0.992 | unsp | BBOV_II000230 | 40 S | KAKHSAKEY | 0.996 | unsp |