

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| BBOV_II002330 | OTHER | 0.999865 | 0.000053 | 0.000082 |
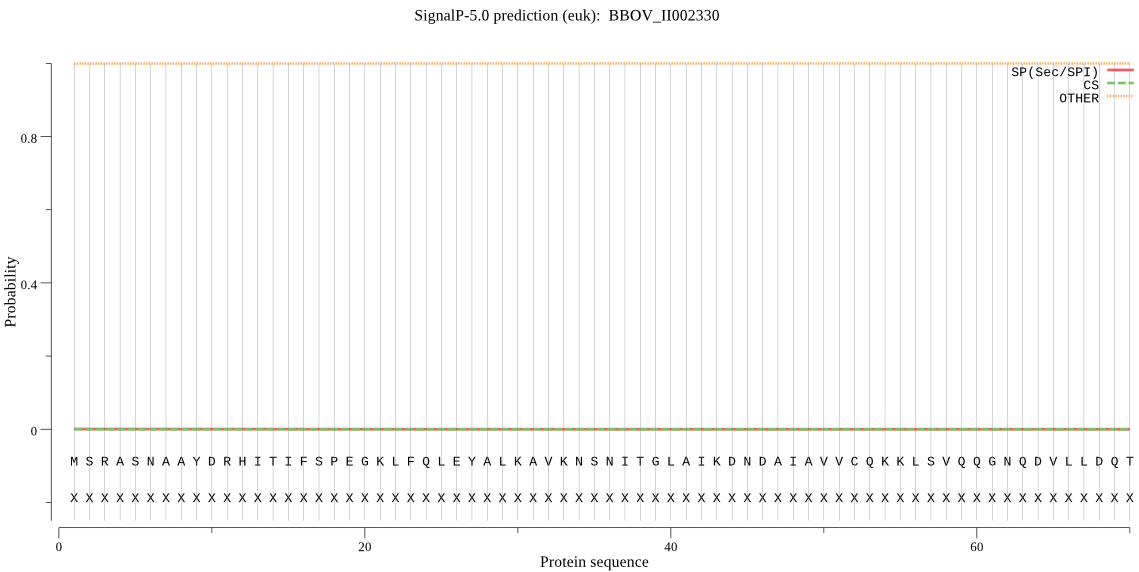
MSRASNAAYDRHITIFSPEGKLFQLEYALKAVKNSNITGLAIKDNDAIAVVCQKKLSVQQ GNQDVLLDQTCVTHLYHITDDIMALLIGLPGDCMSILYKSREIALEYQYKYGCSIPAKVL CEKIADINQVHTQHAYMRLRACTGLIAAIDEELKPVIYKFDASGWYSGYKACGIGTKDQE SENALERILKQRETVDISDKIKSDLQVNTETHHLSGGKETHVTVEGLKALIAIDALSQLN GAKGVEVAVCTIDNPTFRQLSEQEIETYLTYIAETD
| ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|
| BBOV_II002330 | 261 S | FRQLSEQEI | 0.996 | unsp |