

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
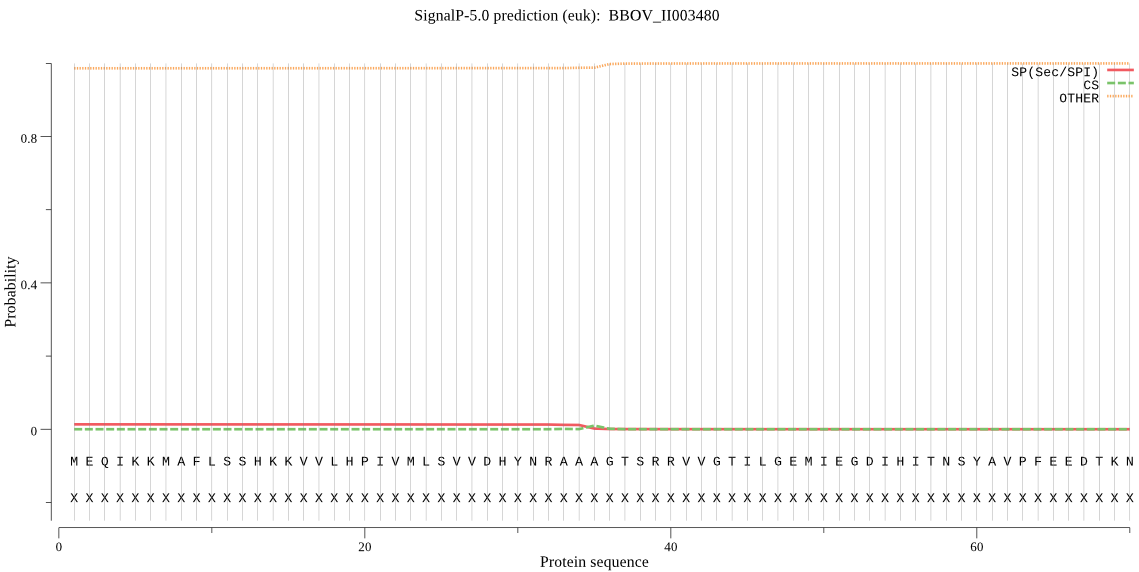
| BBOV_II003480 | OTHER | 0.971295 | 0.007308 | 0.021397 |
MEQIKKMAFLSSHKKVVLHPIVMLSVVDHYNRAAAGTSRRVVGTILGEMIEGDIHITNSY AVPFEEDTKNPLVWFFDHNYHERMFTMFKKINTKEKVLGWYSTGPKCKPADLEIHELYRK YCPNPIYIIVDINQKEEVPIEAYVSVEEPTSDRRFRRTFAHVPSEMGAFEAEEVGLEHLL RDLTNVTTSTLSTKVDNKVLALKSLVLKLAEIVDYLKGVIGGQYNLNPAIVYMLQDILNL FPSVDSDAIKEAFIINMNDTSLAIYLGSILRAILALHNLIDNMTENKRIAKNKKEAAAAQ PKQTSS
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|
| BBOV_II003480 | 145 S | EAYVSVEEP | 0.997 | unsp | BBOV_II003480 | 151 S | EEPTSDRRF | 0.992 | unsp |