

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| BBOV_II004070 | OTHER | 0.999143 | 0.000120 | 0.000737 |
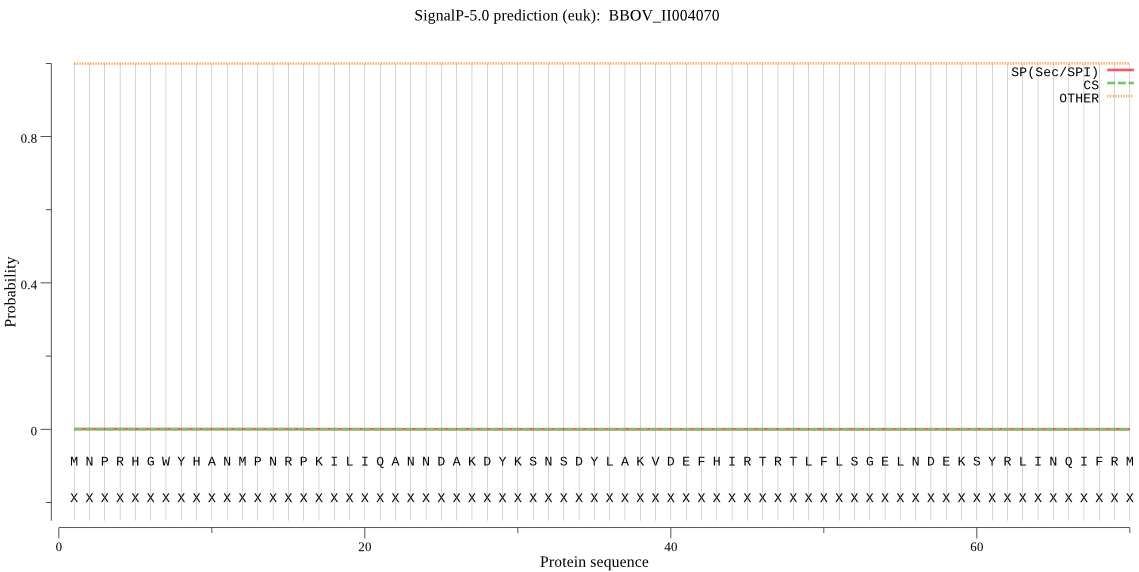
MNPRHGWYHANMPNRPKILIQANNDAKDYKSNSDYLAKVDEFHIRTRTLFLSGELNDEKS YRLINQIFRMNEENPNENIKFYINSPGGSVSAGLAIYDILQSLKMRVETICIGQAASMGA FLLAAGAKGLRSAMPNSRIMIHQPLGGAHGEVTDIKIQANEILHIRQILNAHFSHFTGKS IEQVEQDCERDYYMRPSEAMEYGLIDRICTTRTSHIPIPTAPVIE
| ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|
| BBOV_II004070 | 31 S | KDYKSNSDY | 0.993 | unsp |