

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
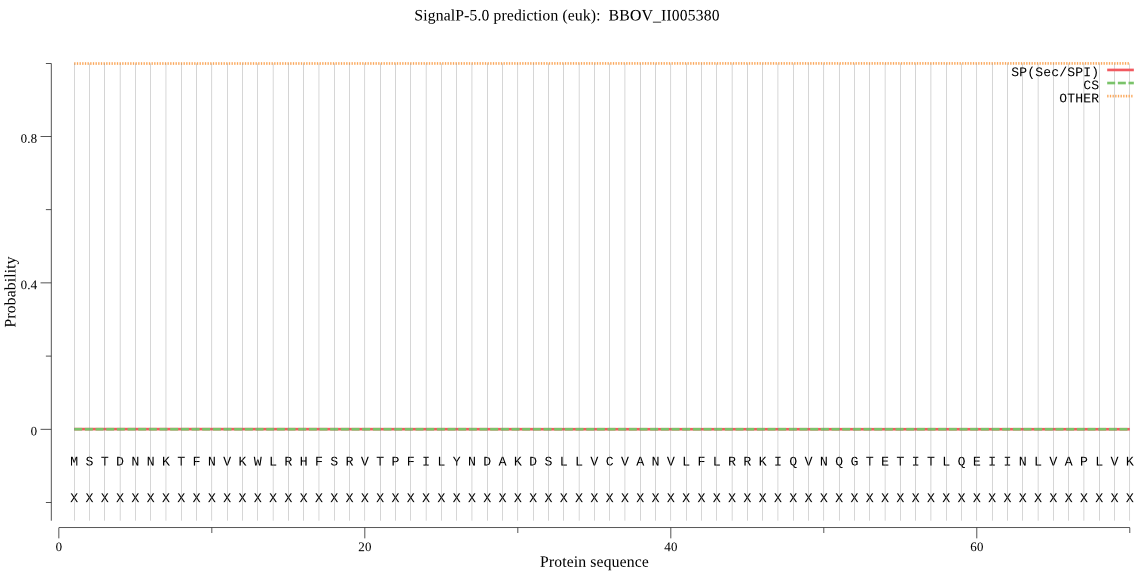
| BBOV_II005380 | OTHER | 0.997479 | 0.001641 | 0.000879 |
MSTDNNKTFNVKWLRHFSRVTPFILYNDAKDSLLVCVANVLFLRRKIQVNQGTETITLQE IINLVAPLVKRVESEWTIETLRSMEKGTKIECPLNAMTVSINQDMMKLLGLLSINLFHGC IAGTRYKSIENFTFDDLRYHMMLHGRGQQMEGVRFEDAEATAADAFMKKYPNRVTEKGLQ MIKAEFRSQENCFHAVYHQNRFQTLTYNDENLYVLMADVIYKSHGCFWEQYDTPAYMDDR FKPYSLAADQVKVITPAGTPRGNEEGVKPIQSRPLEPKVQLVNEAKQLEKPKMEESPPVV PELRIDINEAGKSSSVMNNTAETGINKLDNKGNEEHEPLRIQQTVEENHVNQAGATLMKM MPELAAQNEPAKPNIPAHPGPAISLSIEQPEEPPKIPRRTNEFSQAGSASLLRKNKEQNG QKKSGKCGCQRLRLNDLMEALRGRR
| ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|
| BBOV_II005380 | 128 S | TRYKSIENF | 0.996 | unsp |