

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
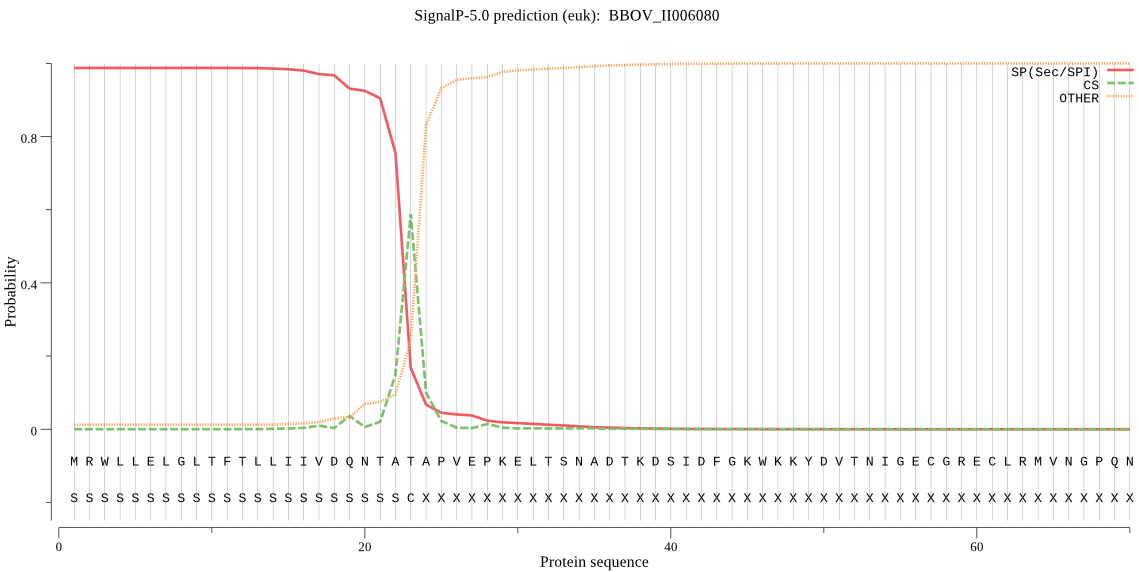
| BBOV_II006080 | SP | 0.008401 | 0.991551 | 0.000048 | CS pos: 23-24. TAT-AP. Pr: 0.4053 |
MRWLLELGLTFTLLIIVDQNTATAPVEPKELTSNADTKDSIDFGKWKKYDVTNIGECGRE CLRMVNGPQNSIPKPIKFIARYTNMNTEQSFLQLSDIVSESTTSTDDIGLVELMSLNMFI VEPPHDYTDEQLESLRKGLTERGIIIENDETAYLTSFDTPKASSTISGSAPVSNHHADNH SAFFNDHMMSDNNIAANSVKNHNESSGRNKRNGGITGAPMKKNRSSKNKRNPANSRKDVQ NEPNIRDDGDGNNKPRRSPKRKINQTEKFQTKMKNHGTNINQGNGQAENKTSDNYSQSTT NIASQNGGVLGPSPDKTNPAIENVKELSDDYSDKQWYITVTQADEAIAKLKTMKRRRVTV CTIDTGVDYNNKSLQHAFEHEGSGESSSEYFGERRYGIDAINETYDPMDIHGHGTGIAGL IAGMKVDGNGITGINPTARLIACKAFDNDLKGRLSDILKCIDYCIKRKAQVQNHSWTLPA ASGALMHAFKILENMNVLSVVSSGNMVPRGPREPNIRLEHVVPATYTTFYSNILSVAGMQ QMTEKNLADRIAQCKSRRNNPESCVVELIDKFELYSKTQYGLHICQVLAPAKNIYTLGLN NTTIVVEGTSYAAGIMSGVSSILLSPIFSNETVGTRIVPHFIQQSVIKLKTCKTTVRWGG YVSASKCLQHIFNNMIIFQTRRLRKWRMARRKLLR
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| BBOV_II006080 | 225 S | KKNRSSKNK | 0.996 | unsp | BBOV_II006080 | 225 S | KKNRSSKNK | 0.996 | unsp | BBOV_II006080 | 225 S | KKNRSSKNK | 0.996 | unsp | BBOV_II006080 | 235 S | NPANSRKDV | 0.997 | unsp | BBOV_II006080 | 258 S | KPRRSPKRK | 0.998 | unsp | BBOV_II006080 | 33 S | KELTSNADT | 0.992 | unsp | BBOV_II006080 | 104 S | ESTTSTDDI | 0.995 | unsp |