

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
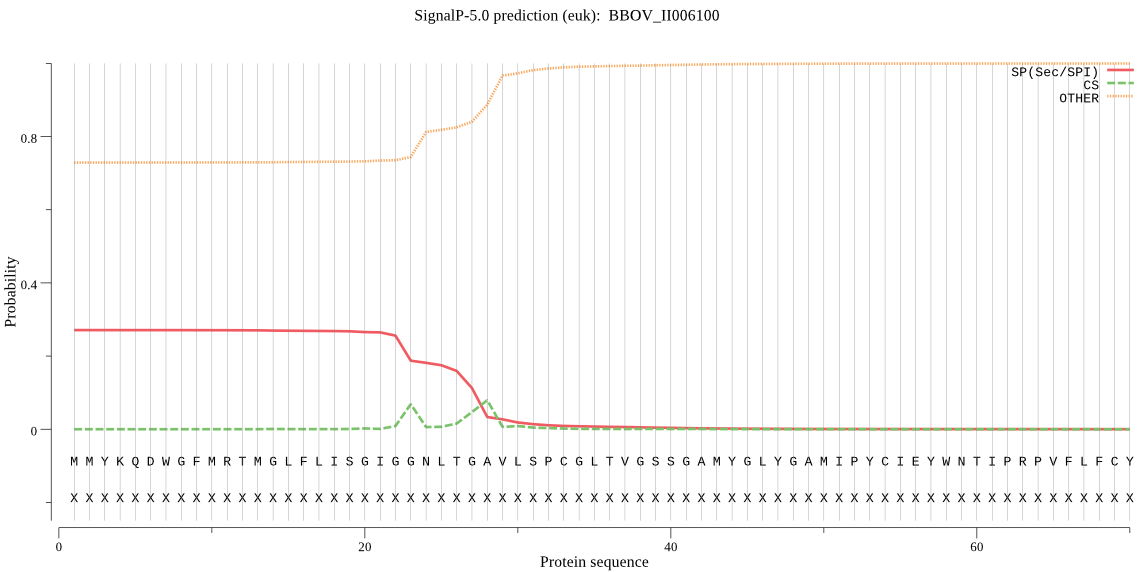
| BBOV_II006100 | OTHER | 0.844467 | 0.145686 | 0.009847 |
MMYKQDWGFMRTMGLFLISGIGGNLTGAVLSPCGLTVGSSGAMYGLYGAMIPYCIEYWNT IPRPVFLFCYNIITLIIGFLMGLAPNVDNYCHIGGCVFGMLWGFATIKSVSSCDKCTIVE RSLLCPLISWALPQKWKAKLRLIIVFKKDRGDRNRQKDMAQKHAVQNGVTPRRIALIKKK FERQGAPPCRMRLREWIFRITSLLMMVLLFTILGLFLSKPELYAKFKPPGEYKLSGWQTC ECCFITNIEKLFSGHRDIPAQYLNRKLFWCFNNVDDAKHFCGDDYAKTAPMDNILKSSHD AALGLINTVTEAIK
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|
| BBOV_II006100 | 111 S | IKSVSSCDK | 0.996 | unsp | BBOV_II006100 | 297 S | NILKSSHDA | 0.991 | unsp |