

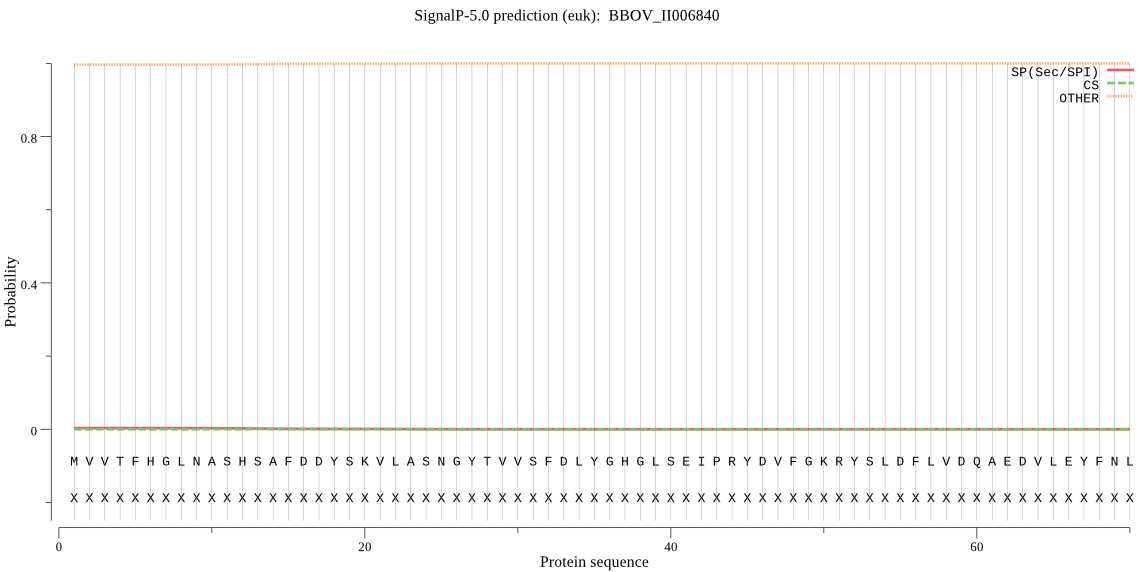
| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| BBOV_II006840 | OTHER | 0.999442 | 0.000435 | 0.000123 |
MVVTFHGLNASHSAFDDYSKVLASNGYTVVSFDLYGHGLSEIPRYDVFGKRYSLDFLVDQ AEDVLEYFNLHDRKITVMGISMGGCIAAAFCDRHPERVERLILISPAGLIPKCPIAAKVV KTLHCLIPCVPLCVCRCCFVAKDKINTQDKLTNHMLWRLYVAPKSSTAMLGIIKRVPLWT AQKLYKRVGAMGKPTLIIFGGKDSVTPPSCAAKLQQLFVNSHVVIFPEACHLASYMIPTA IASTSLAFLSVSTDDKASRYAYWLPFGPNGTYIPRDDRHILNDSEAESSQQFSDDEELLT TPITTHYTETSKIANKSHSRGTLFSKKALVVVNQYEGGDIGTNELLTE