

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
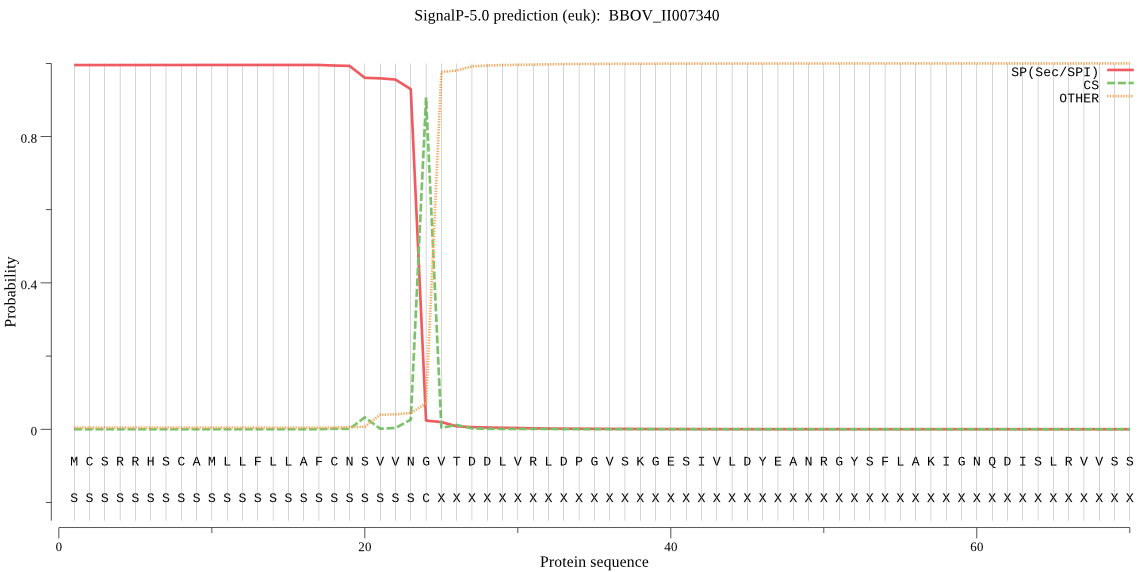
| BBOV_II007340 | SP | 0.000297 | 0.999703 | 0.000001 | CS pos: 24-25. VNG-VT. Pr: 0.7734 |
MCSRRHSCAMLLFLLAFCNSVVNGVTDDLVRLDPGVSKGESIVLDYEANRGYSFLAKIGN QDISLRVVSSLEGLILLCQDTSACSNINLGGICYDPKLSTSAIWCKNSDACIPGEDGYRC VEQSSHEGLQPVTTLDFYSEGIRHRVDTFEGYENVSLRNVGLLVPPIYERVPIKLATNMV VNNSLEFGRDIGGFFGIIGSSASCRGNSMWSDILQRYDGFYMLDLYGDVDTSTVNCIKLG SDVAKRDDILWSEKRQVGGIFTDSATGFTVYGMSMCGTDMFGKTSSNWHVNIDLTSQCLV LPKNFWFSVMAQLPILNSCYRMDGPKLCQLKRGDIEFLPEIQFRLHSQDDSIISIPLENL LLDDTATPRLCIVPQEDTYSPPIFTDRPTIKFGYKVLESLQVVVDTNGYRVGFLPKKPAD TSDAICNPRVDCFGDQQYIPELNECIQPSCSTGVNEIAISDHTSGNSGIYCRETANSP
| ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|
| BBOV_II007340 | 347 S | FRLHSQDDS | 0.994 | unsp |