

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| BBOV_III000270 | OTHER | 0.995847 | 0.003745 | 0.000408 |
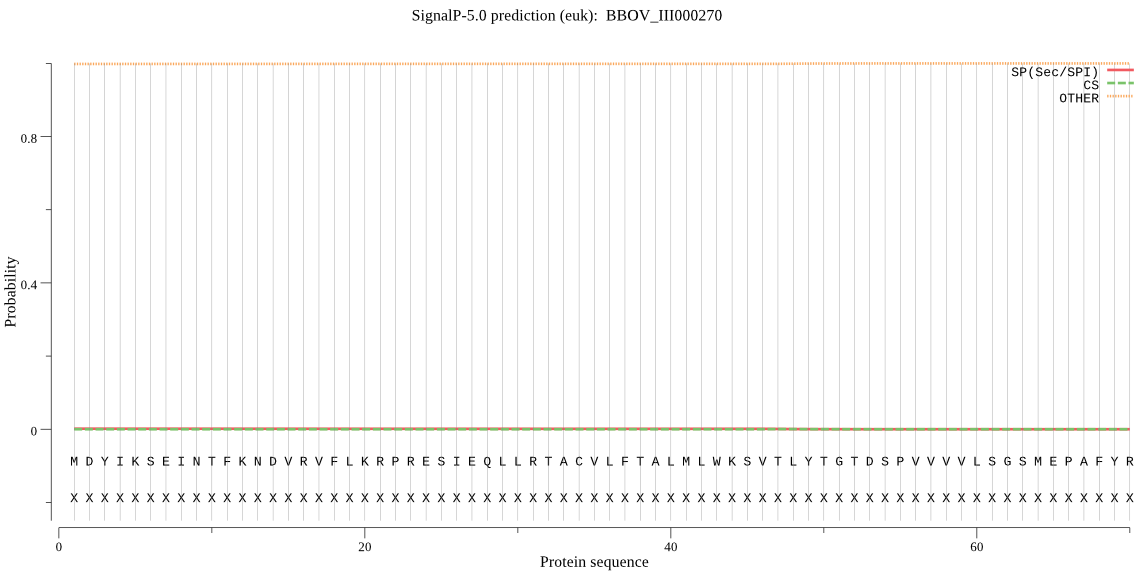
MDYIKSEINTFKNDVRVFLKRPRESIEQLLRTACVLFTALMLWKSVTLYTGTDSPVVVVL SGSMEPAFYRGDILFLMKQEKITAGDIVVFKVEGRDIPIVHRALSLHANGHDINVLTKGD NNEVADRGLYARGQKWLADENILGTVLLCIPKFGLLTIQLNDNPVVKWLLISVLIYLVMT GKG
| ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|
| BBOV_III000270 | 25 S | RPRESIEQL | 0.996 | unsp |