

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| BBOV_III000310 | OTHER | 0.999822 | 0.000058 | 0.000120 |
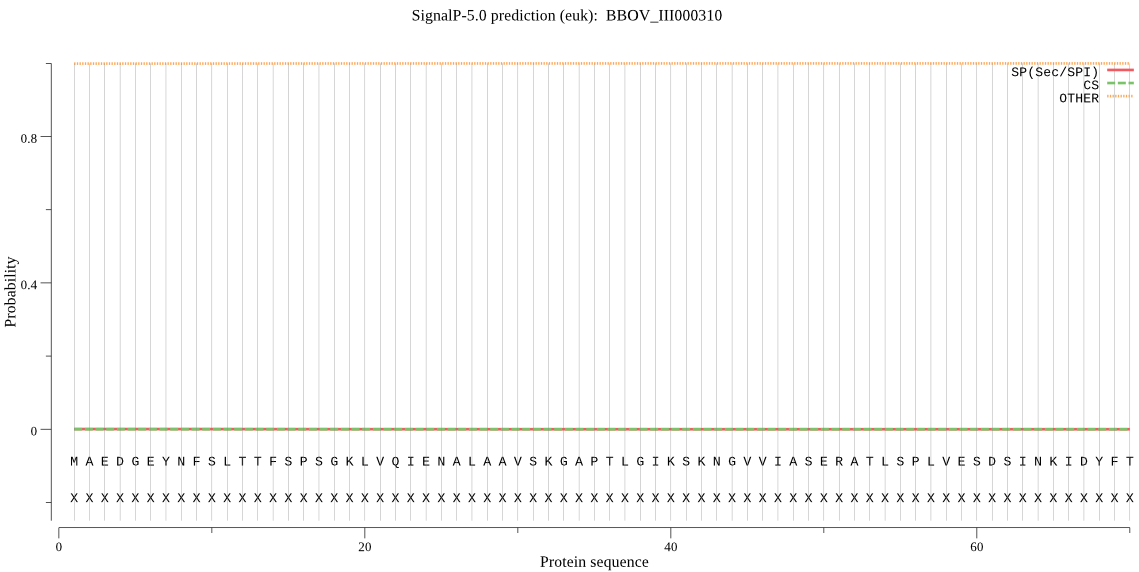
MAEDGEYNFSLTTFSPSGKLVQIENALAAVSKGAPTLGIKSKNGVVIASERATLSPLVES DSINKIDYFTRNIGVVAAGMPADFRIVLKKGRKQAIKYKLLYGDDIPGAELVKEVASIMQ EYTHSGGVRPFGISLLLASYDSDGPQLYQIDPSGAYFGWRATAIGDRMQNNMTFLEKRYN PDLELEDAIHIAILTLKEGFEGEMTTDNIEIGIVGSDGIFKILSKDIVNDYLNELL