

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
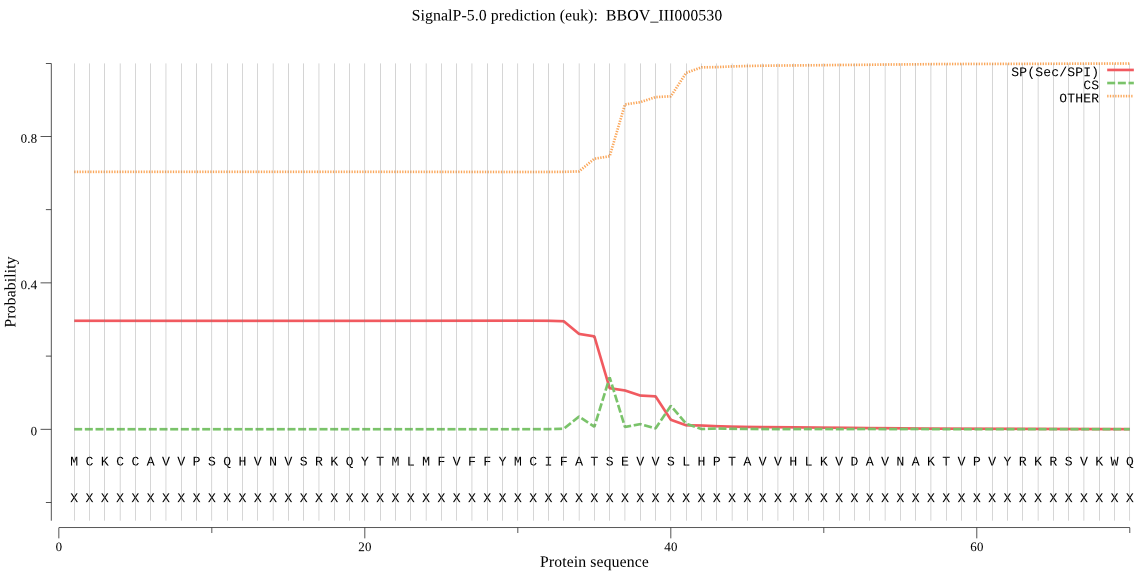
| BBOV_III000530 | SP | 0.425298 | 0.572760 | 0.001942 | CS pos: 36-37. ATS-EV. Pr: 0.5419 |
MCKCCAVVPSQHVNVSRKQYTMLMFVFFYMCIFATSEVVSLHPTAVVHLKVDAVNAKTVP VYRKRSVKWQNPTCAESPCYGQRLLTFIRPKTLIRAPQQVEHASRYETAPLYSSSGIRSY ARCDLQGSPTALAATGNTLAVASVASQINYLRDWVSAKSNAVSRALQNARKTVPKPGIYL YRTCNDIMVKTERYCRTLVTKLHGLYLKAVTLKLKDLISFGKDPVLNIVRVVAMVYALDW WTGLTNIKYNFDLSPFDVFMKGQYYRLFTSLFLHNGVVHLLQNIRSLWAIGYEAADLLGP RRMLTIYLVSGLVANYISYVYYFAYKNAVPAHVFNATQVAVKGLGQMGMSTHHLVEKVVD RIRAKQGALSLPLFMASYLNDTIFHTVDTYLAPLIPMLNRRQALPDCRDVNCAVQQYIKR KCSRYRACGASAAIYGLMGAVCAYYARFGSASDRRRVTEIITSAIGQNIVALFGAKVDHV SHLAGFLTGMCLTMAF
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|
| BBOV_III000530 | 66 S | YRKRSVKWQ | 0.994 | unsp | BBOV_III000530 | 423 S | KRKCSRYRA | 0.995 | unsp |