

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| BBOV_III000740 | SP | 0.487163 | 0.500076 | 0.012761 | CS pos: 29-30. IEA-VN. Pr: 0.9122 |
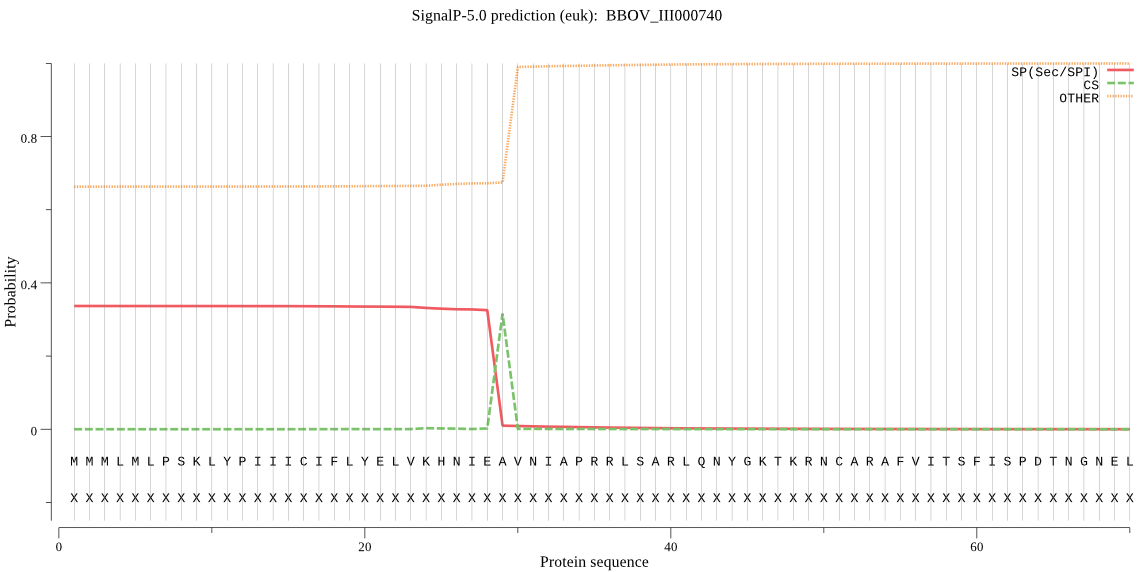
MMMLMLPSKLYPIIICIFLYELVKHNIEAVNIAPRRLSARLQNYGKTKRNCARAFVITSF ISPDTNGNELQERKSLSNTGIVANKRFGKLHGEKSNCSVALSNAMGKCGITISRIIGDTV QELGKRLIENKYYKHGTSLMMLMKQRLSTLSTIEQIALAIIGQCVYAFYLPGVHLIFPYQ IIPTTSGFGVDIGIDSLVSLASSYLLWRSLQKPESHQLTLKPNDTVIIPIVATGLLASFY LSAYAARVVDNVLLLLSAMDWPVDPALQRSVTILLSHLTWVILGVKMLNYVVPLNQKKTL WYTLNSEELWVYKALMGYFVSCGVYNVADLIFNILQAFSKLIHPQLIIKEDITPEFVHVE TSSMIPSIVTALGPCITAPWWEEMLYRVFVFKVINAKLPRNIATCIAALVFAVHHMNPHS IIQLFALGVLWSFIEQGTNNVFISMAIHSLWNTRIMLGTLMGK
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|
| BBOV_III000740 | 75 S | QERKSLSNT | 0.995 | unsp | BBOV_III000740 | 151 S | LSTLSTIEQ | 0.992 | unsp |