

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
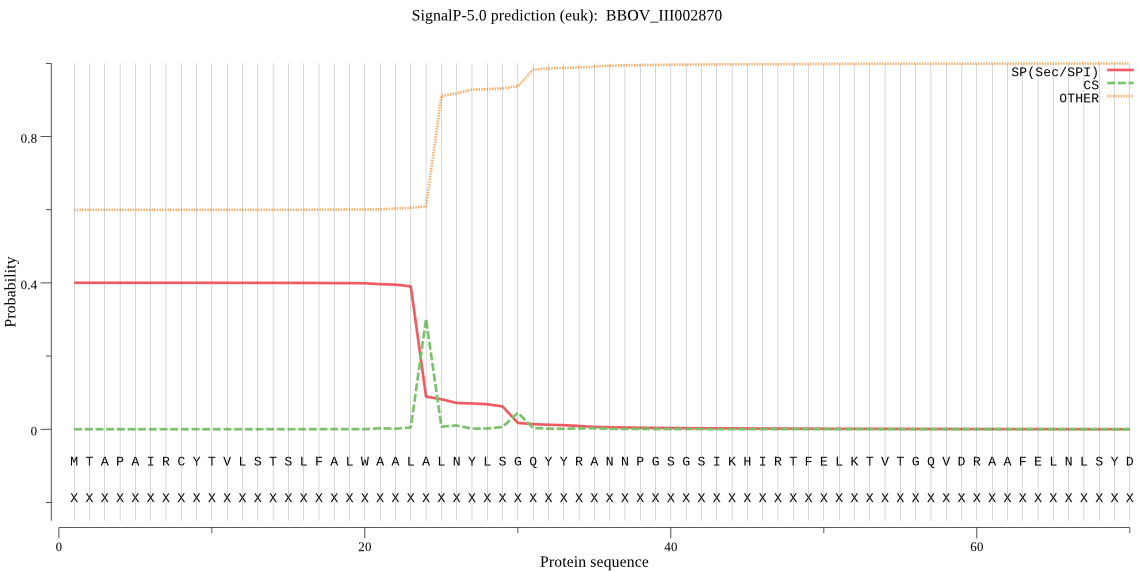
| BBOV_III002870 | SP | 0.253875 | 0.735331 | 0.010795 | CS pos: 30-31. LSG-QY. Pr: 0.4987 |
MTAPAIRCYTVLSTSLFALWAALALNYLSGQYYRANNPGSGSIKHIRTFELKTVTGQVDR AAFELNLSYDLRDVFDWSANVIFLYATVNYETPKHPVNELIIFDKIITSKEEAYEPGADI VSKYYMIDYARSLRKARVTLRLHYCFVPIGGLIKSYQLAESVFTMPSDYVL
| ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|
| BBOV_III002870 | 109 S | KIITSKEEA | 0.994 | unsp |