

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
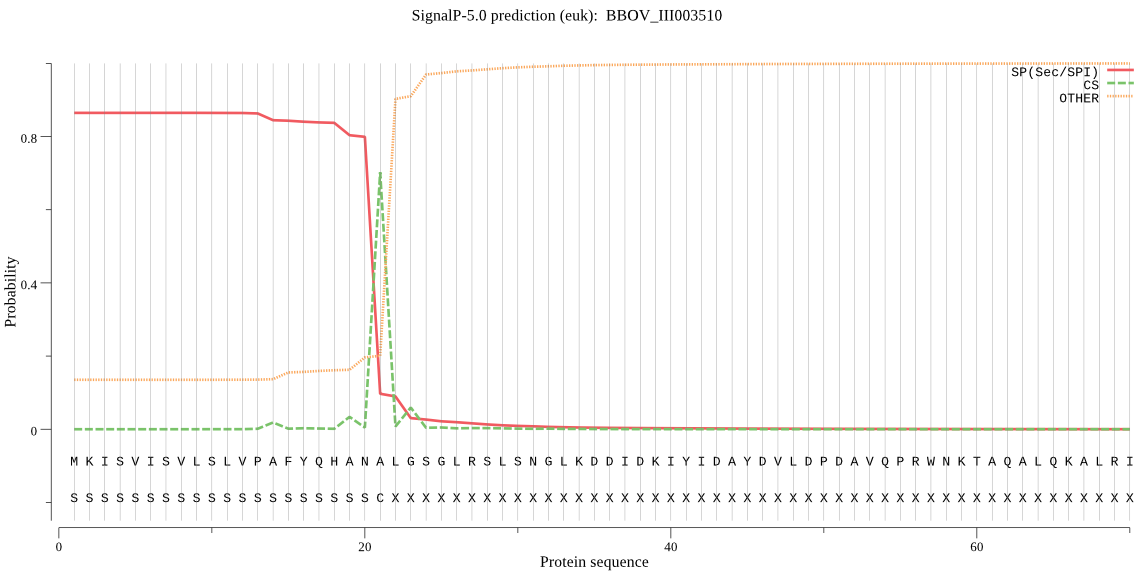
| BBOV_III003510 | SP | 0.093682 | 0.885235 | 0.021084 | CS pos: 21-22. ANA-LG. Pr: 0.8622 |
MKISVISVLSLVPAFYQHANALGSGLRSLSNGLKDDIDKIYIDAYDVLDPDAVQPRWNKT AQALQKALRINYPRMLSSQLFVKPKYEIQLHRFTQVHRKKKESRYQKYTEHDPSFLGHDV ATKQDNIDTFNNTVTRLSQFLLNFENNQYFGEIEVGTPPEKFVVVFDTGSSQLWIPSKEC SSTGCSTHRKFDAKGSSTYKKAPLDAANAYIQYGTGECVLALGSDTVKIGPLEVKNQSIG LATYESDHPFGDLPFDGLVGLGFPDKMYKDTDGMLPLLDNIMQQKLLNRNLVAFYMSKDK TQPGSMSFGSIDPRYVLPGHSPWWFPVVATDFWEIAMEAILIDGKPMKLENKYNAAIDTG SSLISGPSEVVGPLLDRLALSPDCSNLNQLPTISFVFVDMNGRKIKFDMSPDDYVERIEY DDSEFIATKNEPNNDATEQNKNAEIKATENTQCVIGIMTMDVPKPKGPLFVMGVNFINRY MAIFDRDAMAVGLVPSAHKVDEREEQRKLEEEFNIIEGMAM
| ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|
| BBOV_III003510 | 410 S | KFDMSPDDY | 0.997 | unsp |