

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| BBOV_III007870 | OTHER | 0.999650 | 0.000165 | 0.000185 |
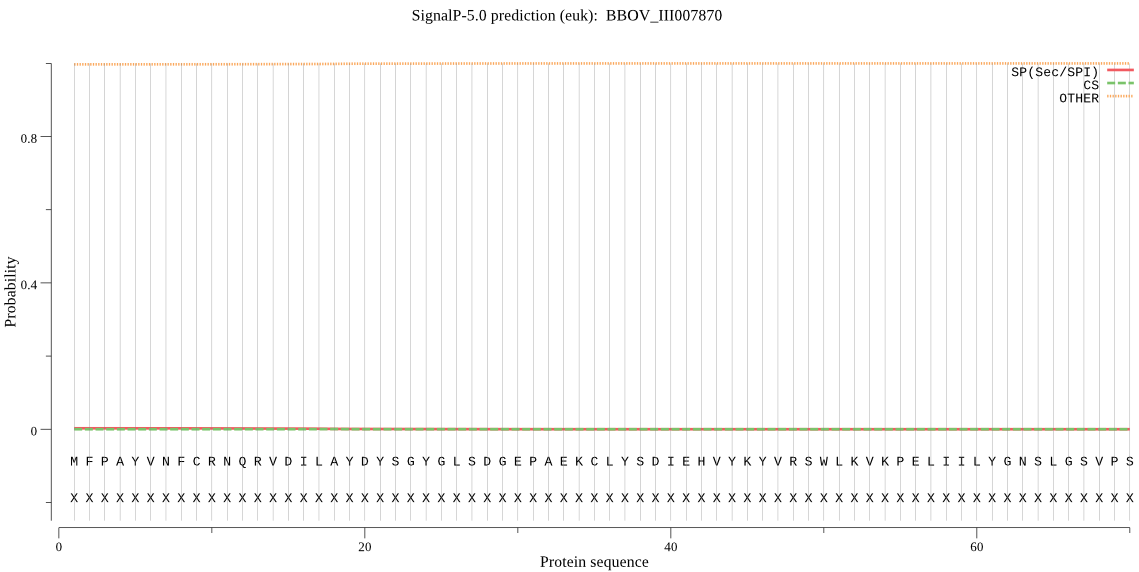
MFPAYVNFCRNQRVDILAYDYSGYGLSDGEPAEKCLYSDIEHVYKYVRSWLKVKPELIIL YGNSLGSVPSSYLASMPEKYPIGGLILDAPLSSAIRLQVGYVKKTPRFDAFANIEYLKSK ALYPTLVIHGTSDGIIPIEHARDLAFVVEVRHAELMPEGNYTLQFDMESSKESCDDSVPV FSRPGIELIKQPEDLLRTWWVPGAGHNNIQLDYPEAYVRTIEVFFKLCTTWRINRKG
| ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|
| BBOV_III007870 | 173 S | SSKESCDDS | 0.996 | unsp |