

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| BBOV_III009430 | OTHER | 0.513831 | 0.484406 | 0.001763 |
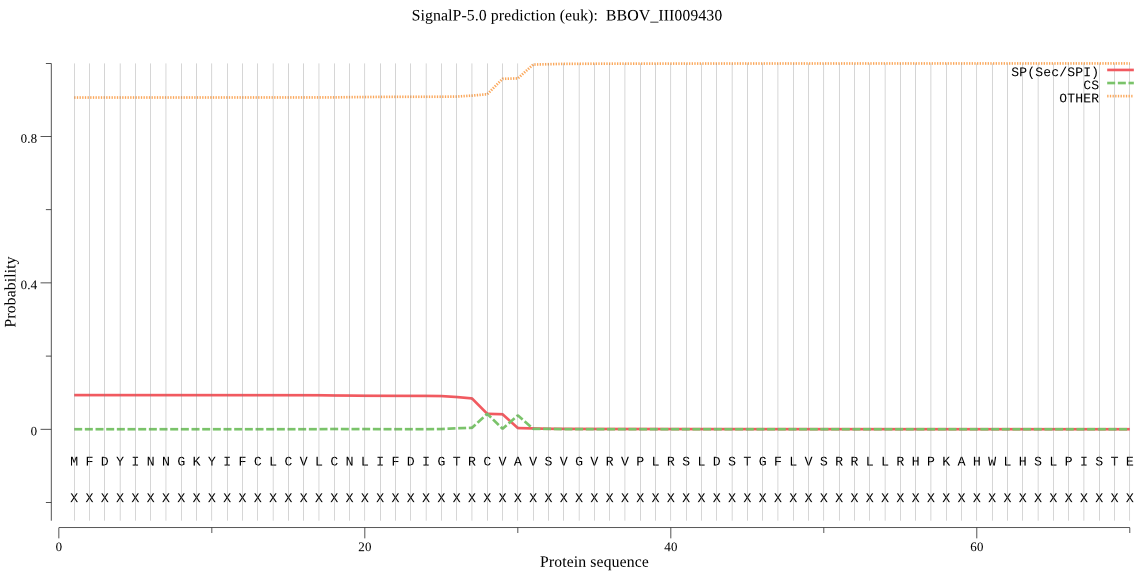
MFDYINNGKYIFCLCVLCNLIFDIGTRCVAVSVGVRVPLRSLDSTGFLVSRRLLRHPKAH WLHSLPISTEEDVRCSYTTQLESCLEQLKSDCPANGSNYYILAIETSCDDCCAAVVSSNG DVVSEERASNPDSLIKFGGIKPDESYRFHLDNIDRIMNEVVSKAKLKFEDIGYIVATRGP GMRICLNAGYDAAERISKTYSIPLIGENHLAGHCLSPFIKGHQLRMTHDRGSVASEELKY PYLSLLLSGGHSQIYVVESPYQYHMLVDTMDHYAGNVLYKCAKELGLPIDTGGGPSIEEA ARKRQGRPMFRMTEPCKGMSFTSFCFSGIQTQLRSMVSKIRQDLGEDALSEDPKLVNHLA YTCQEVTFNQVIRQLDKALDICETLFGISQIVVVGGRSGFIENKFF
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| BBOV_III009430 | 145 S | KPDESYRFH | 0.991 | unsp | BBOV_III009430 | 145 S | KPDESYRFH | 0.991 | unsp | BBOV_III009430 | 145 S | KPDESYRFH | 0.991 | unsp | BBOV_III009430 | 232 S | HDRGSVASE | 0.994 | unsp | BBOV_III009430 | 296 S | GGGPSIEEA | 0.995 | unsp | BBOV_III009430 | 68 S | SLPISTEED | 0.994 | unsp | BBOV_III009430 | 129 S | EERASNPDS | 0.997 | unsp |