

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| BBOV_III010070 | OTHER | 0.999634 | 0.000358 | 0.000008 |
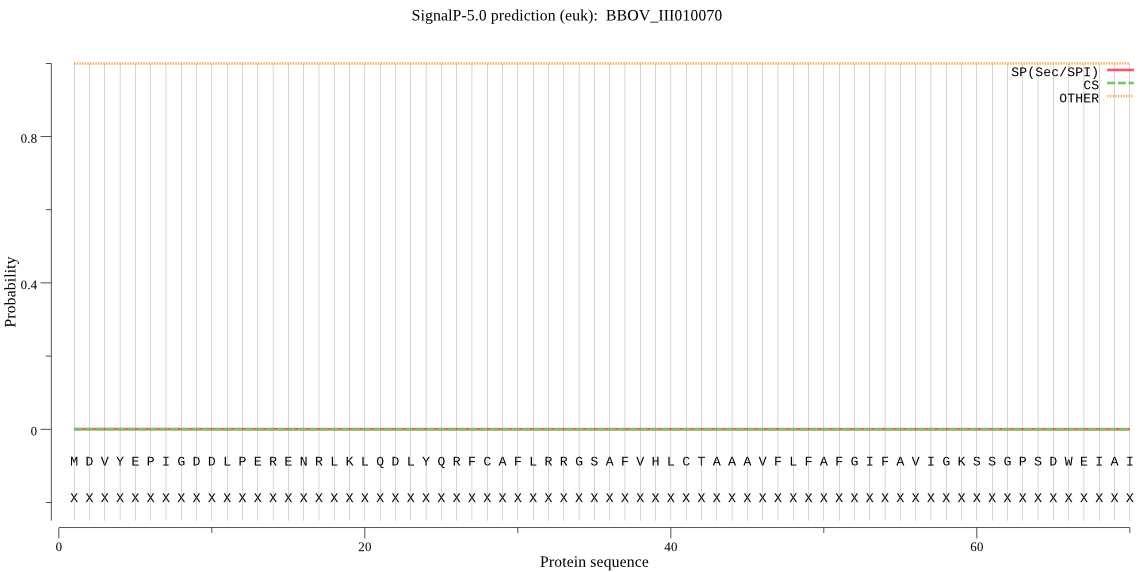
MDVYEPIGDDLPERENRLKLQDLYQRFCAFLRRGSAFVHLCTAAAVFLFAFGIFAVIGKS SGPSDWEIAIRNELLHRNFENHATVLGEYSEDPAAIIVEALKRDQVLIGASTAVELYIQF NDFNRDFKRHDNSISEKIERFATFYRNVTRIREFNMNVHKTYTMKINQFADMTPEQFMSL QGTRASKIRVSKGIPDSQVAAVGNQKGPNLKSEVRQTGNRFADISPEDFIDLRKDNYMTP VKDQGNCGSCWAFSLIGVAEPFFKHKRDIDVVLSEQNLVDCVKECHGCDYGNSYFAYEYI RDHGVYRLASYPYTAKSGPCVEPLNEPRLTISRFGLSENPDLPQLLKQYGPLTVYVAVNV DWQFYSSGILDSCADEINHAVVLAGVGQDDDGPFWLIKNSWGTSWGEEGYVRLARGSSAF DNECGLAHMALYASA
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|
| BBOV_III010070 | 225 S | FADISPEDF | 0.997 | unsp | BBOV_III010070 | 418 S | ARGSSAFDN | 0.992 | unsp |