

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| BBOV_IV000300 | OTHER | 0.998054 | 0.000936 | 0.001010 |
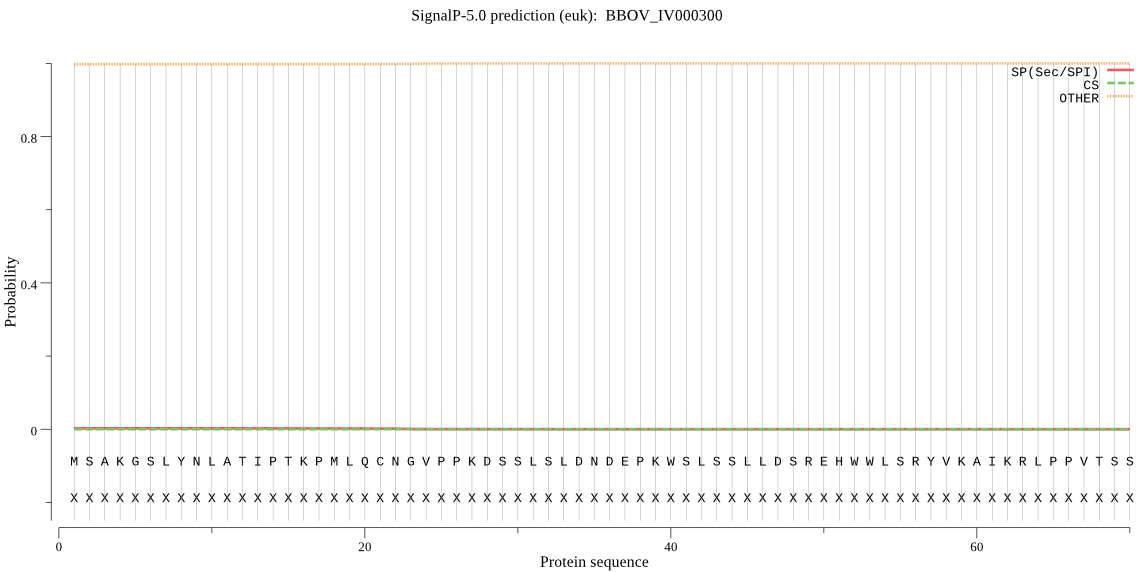
MSAKGSLYNLATIPTKPMLQCNGVPPKDSSLSLDNDEPKWSLSSLLDSREHWWLSRYVKA IKRLPPVTSSYITLSTIMALLSWGFNDNYPLEILQFDLKSFKSGKIWRILTPFLNFGQLW LAHIFMAQSVALYMASVEVSHCAKPEKFLEFMTFGTMCISLYGLADSLFGKQENTMGSAA YHLHTYVLYYWSRLNEGTVVNCFDLFNLPAEMVPFIFVLQNYLLYKEIYATDIAAIAAAH FYFYNLSNTKAIWPFSLLRKIGFKKLYERFNNEISR