

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| BBOV_IV001950 | SP | 0.047819 | 0.883578 | 0.068604 | CS pos: 17-18. YNA-NV. Pr: 0.4576 |
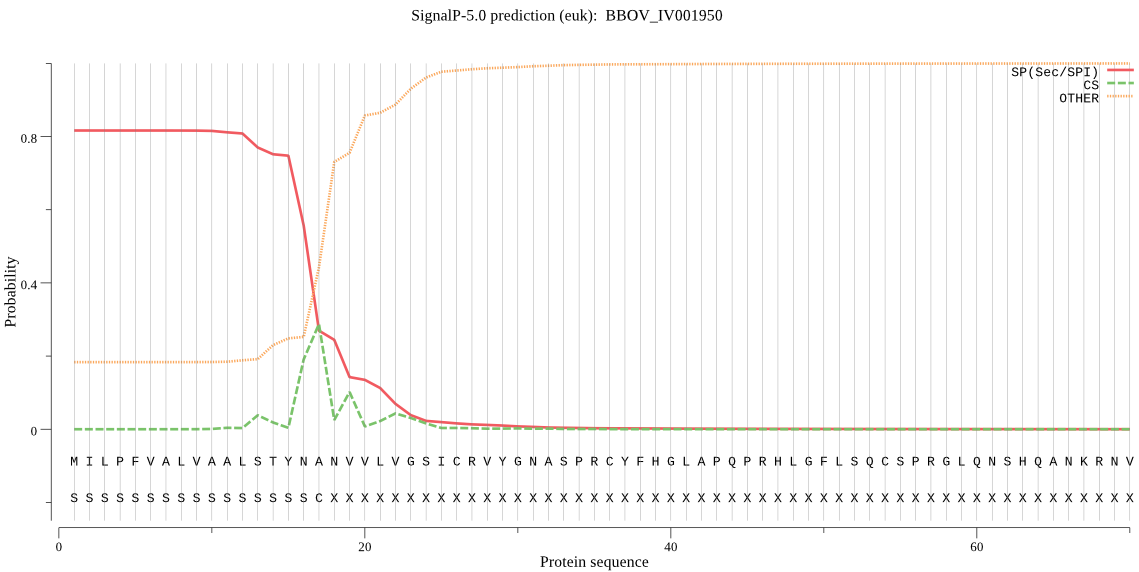
MILPFVALVAALSTYNANVVLVGSICRVYGNASPRCYFHGLAPQPRHLGFLSQCSPRGLQ NSHQANKRNVNAIYSQKHILDKERYIRRPKPEGFGIKKKKTPEPKSNRTKEIVDDFAKQY PSASIKELWEHYEFTGPIKKGIVTGRLLPGKNIKRPNYYASGLPKYVDYPRDVDYRGEHD PRGTIKTDSEIAGIRYACRIAREVLDSVSPLIVEGMFTDYIDRHIHRMCRLKKVYPSPLN YHGFPKSVCTSINEIVCHGIPDSTVLAAGDIINVDVTVYADGFHGDVSETFMVLPAEDPK KLASMNHDIGMYERNKLRYNAKFHDNLWDSGATVMTELNILQCMDTAVRRLGPQDGWEEW FDMLYTGDPGRRPIFGTKNNDDLDPVAKRIMPDIEAWYTANEAVTYKGRVVGVVESQIPT YIAPPGQPKRYATLENPEVIHHIFNRVDPDRRKKPYLFTHTFKEDIELMQITHDAMMKAI AICAPGVPISRIGDVISDHVEQYGFRVVPNLVGHGIGRNFHENPIIEHTRNQSEVVMEPG MVFTIEPIVTRSMDCEVITWPDGWTMATSDARKTAQFEHTVLITDHGHEILTQRLPSSPP LIWNTDLQVMY