

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| BBOV_IV003850 | SP | 0.365511 | 0.631586 | 0.002903 | CS pos: 22-23. FIA-TF. Pr: 0.4635 |
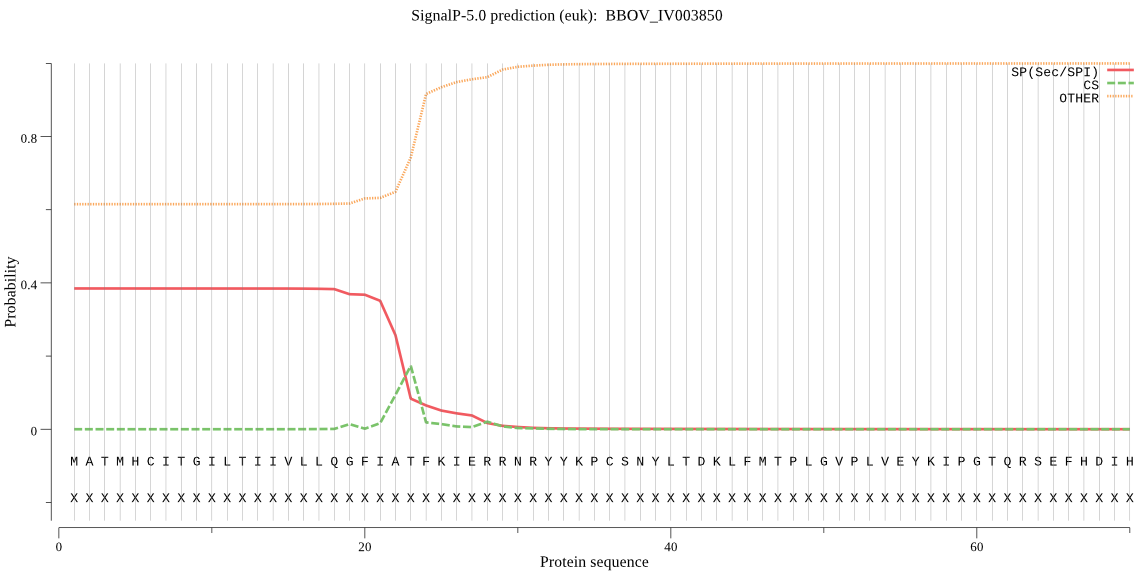
MATMHCITGILTIIVLLQGFIATFKIERRNRYYKPCSNYLTDKLFMTPLGVPLVEYKIPG TQRSEFHDIHTRLYRERILFVSQSLDESYTNMIIGTLLCLDSDSNEKPITLYINSQGGPS TAGITLYDTIKHLKSKIATINVGFCSATASLLLASGTPGMRSALPHSRVVMHQPAGTLEG SAEQIRSQAQYMLNMKRILQKLYSQVTNQPLDNIAKDLEREQFMSAEESLQYGLVDRIIR TDHAK
| ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|
| BBOV_IV003850 | 225 S | EQFMSAEES | 0.993 | unsp |