

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| BBOV_IV005690 | OTHER | 0.998954 | 0.000248 | 0.000798 |
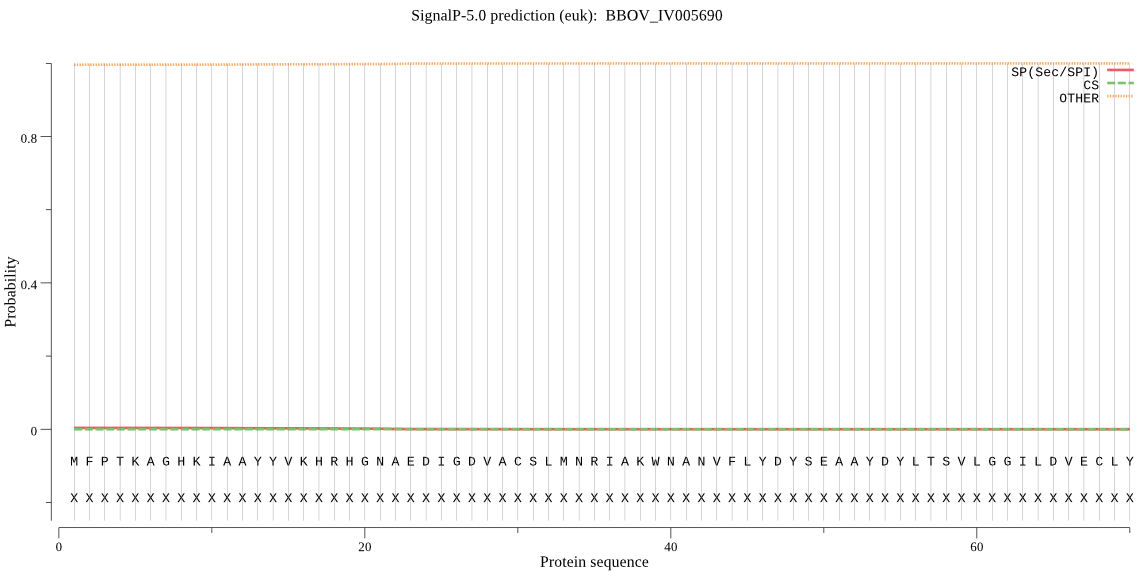
MFPTKAGHKIAAYYVKHRHGNAEDIGDVACSLMNRIAKWNANVFLYDYSEAAYDYLTSVL GGILDVECLYIIGVNPHTIVAYGRSIGSGPTVHIALKRSVLGVVLQSPISSVYKVKVYRL PCTIPGDMFRNEDKVDRINVPTLILHGTKDNVVPISISQSMALTMQRVYGRWINGAGHDD MDTTFALYVDQALQEFYDIICPRTTISNIPKDTAT