

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| BBOV_IV007800 | OTHER | 0.999730 | 0.000257 | 0.000013 |
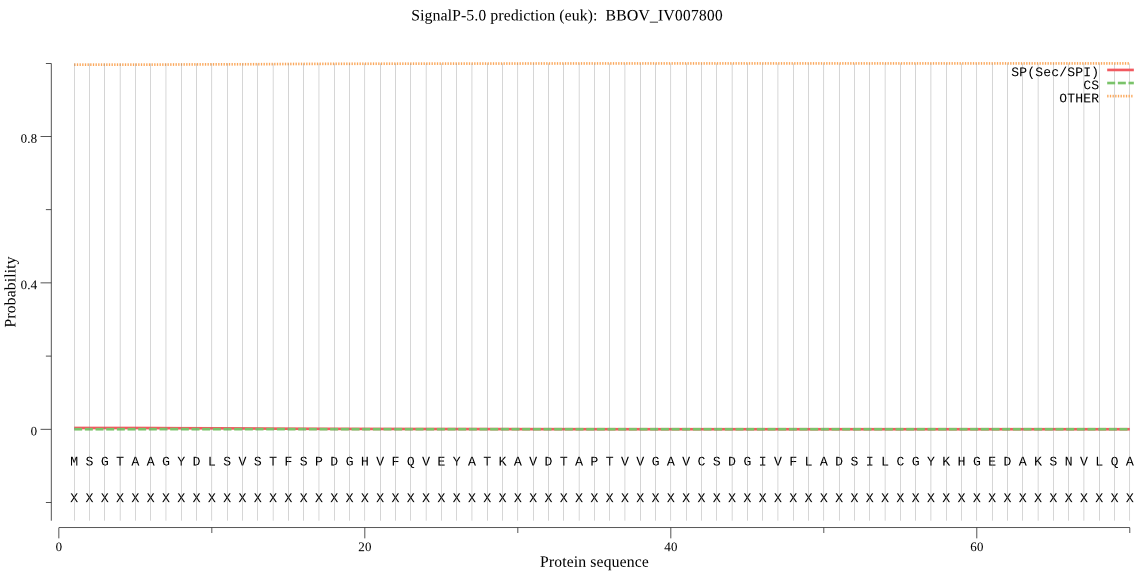
MSGTAAGYDLSVSTFSPDGHVFQVEYATKAVDTAPTVVGAVCSDGIVFLADSILCGYKHG EDAKSNVLQARPTIRLYAIDEGVGCAVTGMIPDAQCIVRHAKSECKSFFAEYGVKIPVSL LAERVALFVHAYTLYWHVRPFGASMILSGIDSNGEKSLYCIEPSGACYKYAGMAIGKCKH LVKTEMEKFILSGMSCRDALKELSLAIMIGRDEDSGKTNDLQMAWICDESGGEFQMIPED IAAETKADASRRHAAIHGQ
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|
| BBOV_IV007800 | 195 S | LSGMSCRDA | 0.996 | unsp | BBOV_IV007800 | 215 S | RDEDSGKTN | 0.993 | unsp |