

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
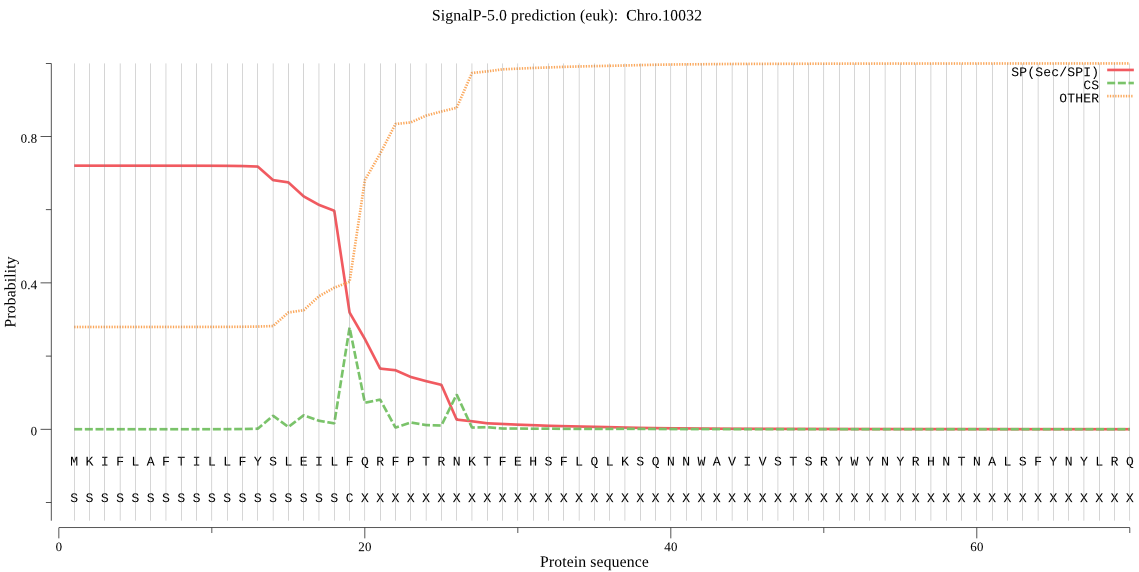
| Chro.10032 | SP | 0.139363 | 0.860165 | 0.000471 | CS pos: 20-21. LFQ-RF. Pr: 0.3825 |
MKIFLAFTILLFYSLEILFQRFPTRNKTFEHSFLQLKSQNNWAVIVSTSRYWYNYRHNTN ALSFYNYLRQNGFRDDRIILMLAENIPCNTRNSIPGGVYSEDFDFFYNLNNHTQTMECAD VDYREDEVTVSNFIKVLTNKHDDSVPNKKRLLSDEDSNIFIFLTGHGGDGFLKFQDFEEM TSFELANAIKEMKAHKRFKKIFIISETCQASTLHNHLDFEDVYAIGCSSLGESSYSKHYK VEIGVASIDRFTHFSLADFKNLNRNKLMPIASLIGKYSVFQLKSTPQLKYKSGKTDIKNV YVNEFFFPNIEKLFSLNIGNLILNHKNIKEPIKRVLNEGVSKYLNCSFLYHTLKRYNDLD LIDPLSNCHFKNTLKKAIIFSQKRYVFNSKFSSKKMIDNKSLKLIKAIFGLTFILILVFI LSYYSL
| ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|
| Chro.10032 | 153 S | KRLLSDEDS | 0.998 | unsp |