

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
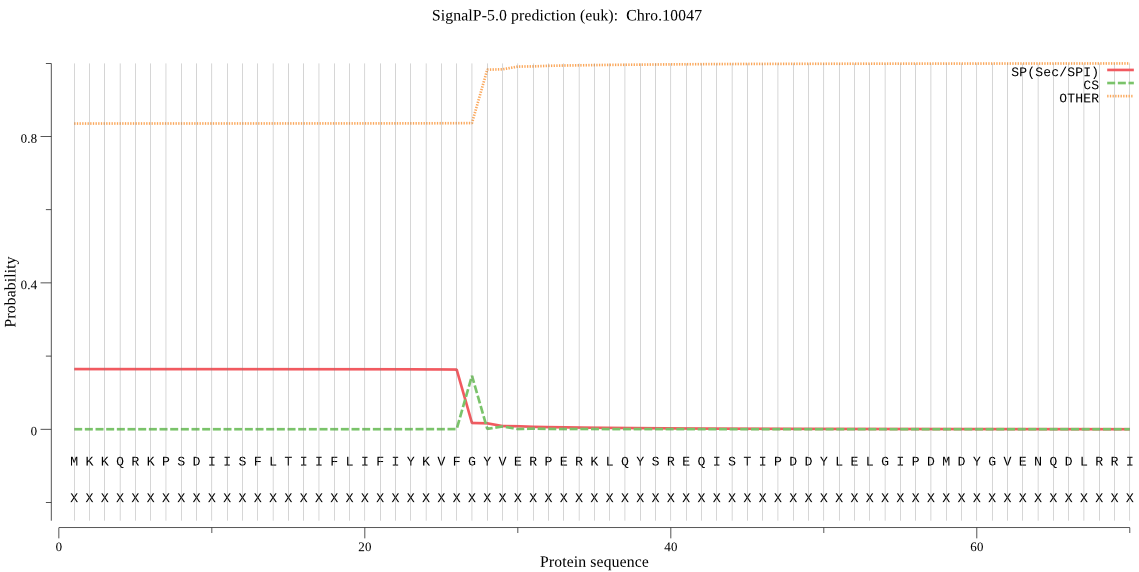
| Chro.10047 | SP | 0.348988 | 0.644598 | 0.006414 | CS pos: 27-28. VFG-YV. Pr: 0.9843 |
MKKQRKPSDIISFLTIIFLIFIYKVFGYVERPERKLQYSREQISTIPDDYLELGIPDMDY GVENQDLRRILYSLNRGSLRSFPTLNEAMIILDNLASQFGPNFMRKHKIGTSFEGRPIDA YRVGFFSKMDGSYSEDSYFYINQGKKPAFLLTSMHHSREPAGLTTGIYFITKLMEDAIYK QDPASNFLLSNIDIWYVPFVNPDGYAAIERTRNYGIRKNQRKTCNSGRSDEDGVDINRNY DFNFKNSLVSKCDPQEYSGEYPFSEPETRAIRDLVNNVKSFVTAVNLHTFGDLWTIPWNC CKNKDLNENIASIYNELKYEILISSPSCIIRTLMHMKGPFFVNSAKKGFDSTFFTKSRNE FENASNYCFSSAARNPTMDYEASGEADDYLLAVHNIISLSPEIGSEYFGFYPPQSEIFPI AKKYYPQILAVASKSTIELSISASLHLQMHSEVNHGKLEFSMFNSGLSSICSNITYETHK NDSKGTSECHTIFTWELLSSNCKQDSKFDPGKTKFGEYSAGFRYRTSNFKELNSTVS
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|
| Chro.10047 | 229 S | NSGRSDEDG | 0.997 | unsp | Chro.10047 | 250 S | NSLVSKCDP | 0.994 | unsp |