

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| Chro.10193 | OTHER | 0.999170 | 0.000066 | 0.000763 |
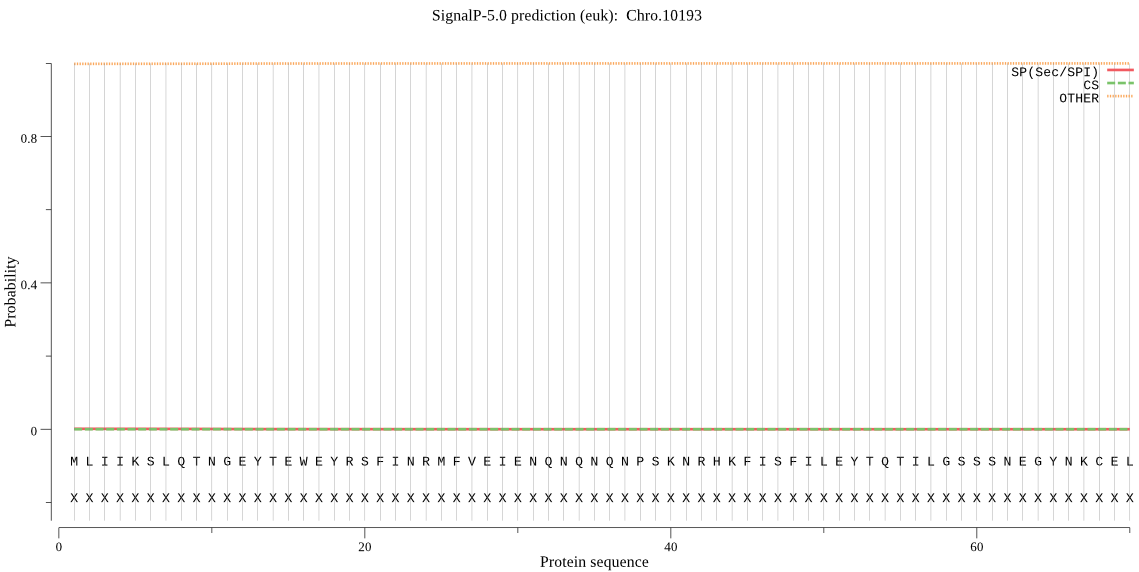
MLIIKSLQTNGEYTEWEYRSFINRMFVEIENQNQNQNPSKNRHKFISFILEYTQTILGSS SNEGYNKCELFNSWAYKLLHRQSIIAYLQGNISKNKSLYLIEKFVLNSKILSLNDKYSMK KKIHKLTRPIDIALINPVVEDINNTVLAFYQFGVPSFEEKLHLMALQPIIQEYIYDNLRT NKQLGYIIFANIIPISSTRLLVVGVEGDNNNSVEKIESIIRNTLYEFSTRKLGNMESHMF EDIKNSLIQEAKSIGNSFNQKLNHYWDEIRYVGDLSESFNLQRAIDYINNKMTIEHLYNT FNKLINSEERTRSHSTIKAIYYSKTSPEFKKQEFIDKYMNSALSIARMSLKEDDFY
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Chro.10193 | 326 S | YSKTSPEFK | 0.996 | unsp | Chro.10193 | 326 S | YSKTSPEFK | 0.996 | unsp | Chro.10193 | 326 S | YSKTSPEFK | 0.996 | unsp | Chro.10193 | 349 S | IARMSLKED | 0.998 | unsp | Chro.10193 | 253 S | QEAKSIGNS | 0.994 | unsp | Chro.10193 | 315 S | TRSHSTIKA | 0.997 | unsp |