

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| Chro.10321 | SP | 0.011084 | 0.988556 | 0.000360 | CS pos: 24-25. CHG-NK. Pr: 0.8816 |
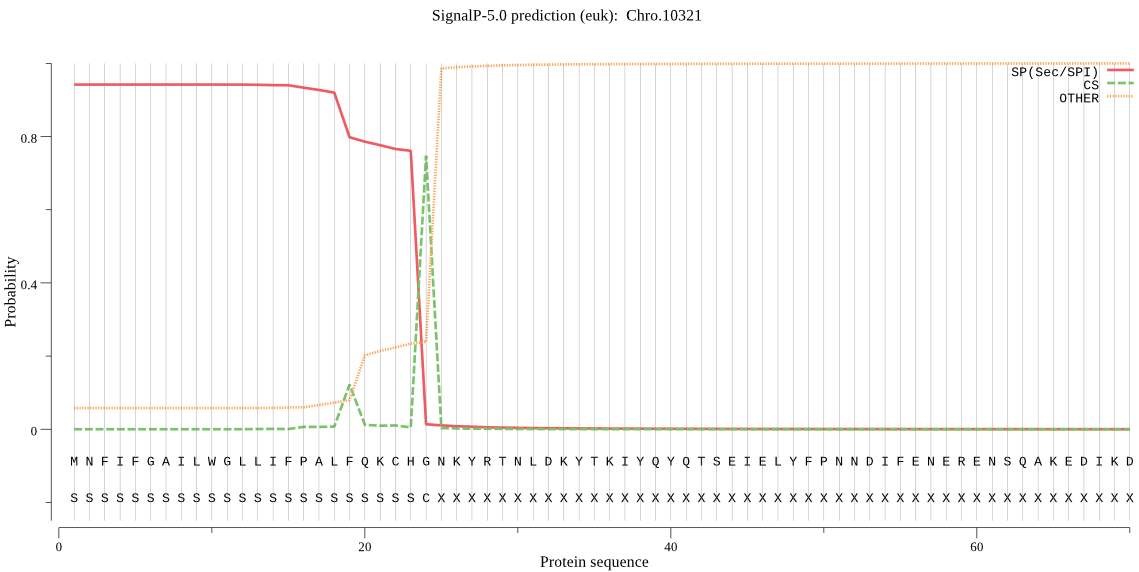
MNFIFGAILWGLLIFPALFQKCHGNKYRTNLDKYTKIYQYQTSEIELYFPNNDIFENERE NSQAKEDIKDLKSTEVSDIRYSFMKLGGKEFLCGFEGHGKNDSLNLSEETNQSWDNMNSK IKNYLKKSKISWLIERCFIYTKKKDINNGIHVQVDTFEICVGVSVRHKRQIIDKETSDNI LESQFNIVGDYKPTEDTFYENGTIIHYYNPAIENLKGDSGYSASIEFKCSYNKSGISQVT EDVNKEFKPVIKVDFLSPSFCDWRVDESKNITSLHKLESLLLPLENRCQNFTDSRFWNYE LCNLYAVSQFKKDASTQELRLFNLGIHPNTAQILNITDDLEEYLSKNLISNIVNTIKPGR VSLLENITTKIEPRNINELISGKYVHSKYVITVMLLNGTHCPENNQKRSIKLVFECPDNF ESISDYFRIVNVIEDSTCNYEMLIISPVICSHPLLMPPPIYKHGKVKCIPKNLIINKLIE DESTEHLFESQGTRSESGLVSISQIGNQFEAVNIEYINSIKNGSGTQHFFTLSDSPRIRN SNLTSPKFLVGQIVQHLWWNYYGVVVGWDWELNAPKQWADNIYQKYPPESRKKPHYLLLV HQKETLFNNSSPIESIPSNLTHSYIPEIALKRVPNLDSDNNFSSKNIIINKYLNTYFSYW SEIHQRFIPNTNSTLWRIYPKDFDKLNERDEL
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Chro.10321 | 590 S | YPPESRKKP | 0.994 | unsp | Chro.10321 | 590 S | YPPESRKKP | 0.994 | unsp | Chro.10321 | 590 S | YPPESRKKP | 0.994 | unsp | Chro.10321 | 73 S | KDLKSTEVS | 0.996 | unsp | Chro.10321 | 315 S | KKDASTQEL | 0.993 | unsp |