

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
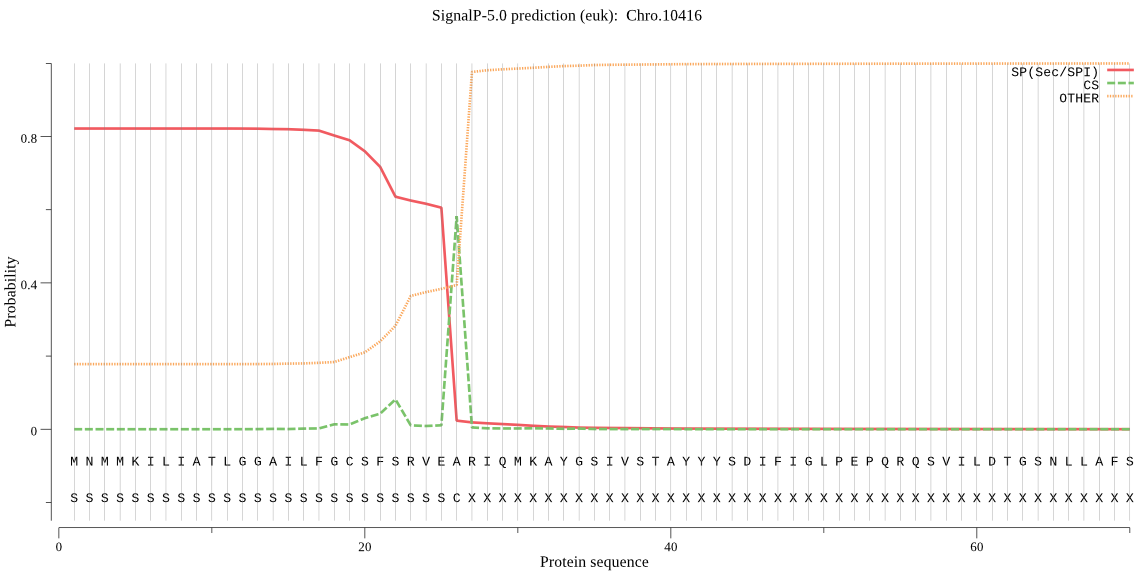
| Chro.10416 | SP | 0.007712 | 0.991775 | 0.000513 | CS pos: 26-27. VEA-RI. Pr: 0.5720 |
MNMMKILIATLGGAILFGCSFSRVEARIQMKAYGSIVSTAYYYSDIFIGLPEPQRQSVIL DTGSNLLAFSSTQCQQCGTHLDAYYDPFKSITKREVPCHSYCKVCVNNKKQCAYTIHYLE GSSLSGSYFEDFVAIRNEKGINSEPSPYVVGLSTIFGGITHETNLFFTQAASGILGLAYT ASSQERAPLFQTWTKRSKYAKDAILSLCFSSEGGMISFGGYNSEYWVLGSNDKSFRSTSE ANLFERALGYSFLSSSSNSQSRSNSRGNNLDSKIGWTPLSILNGNYYVQLTKVSVYQTKL TLHKDSSSGNTKPIPLVIDSGTTLSYFPEHIFIQILNVINQRIAETENKSTFRSLIEAGS KFLEYTGLKSSSSQNELELGIKQISIFPGEVPGAALLRKRVYRRLNNIEDIVNTFNSTVH PENFSDEDIESSNNQLNNSNEVSNHFNQTTTSTTTSTINTLFSSQEFDIDSSVSRIMLET SKGERCWKLRDPNEMSRFPTITLGFPGLKVEWEPAQYLYKKYRNTYCLGFDSDKSFLVLG ASFFINKDVIIDVKNSRASFVKSNCPQIAHARRTSTSEMSQLLKDASSSSPESIMKSEVV VFDSNTKHINIENGNSNNNNNNNNSQSLNLSSGTSSKNPPNYRHSADNDTISESHWEPKN LTSPSSLSSYLSQSAVQYIGVTNHSVDYYNQSINYENLGNNDMLILYLKQQLHDIDDIAD NTGNPQIQKNLQDANNSPSDSKNENQDANSDNKDSSSRYSNSQPTIIHWLIILLTALFSC YLFNGVDQKDAKTE
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Chro.10416 | 575 S | ARRTSTSEM | 0.997 | unsp | Chro.10416 | 575 S | ARRTSTSEM | 0.997 | unsp | Chro.10416 | 575 S | ARRTSTSEM | 0.997 | unsp | Chro.10416 | 590 S | ASSSSPESI | 0.995 | unsp | Chro.10416 | 635 S | SSGTSSKNP | 0.993 | unsp | Chro.10416 | 737 S | DANNSPSDS | 0.996 | unsp | Chro.10416 | 183 S | YTASSQERA | 0.996 | unsp | Chro.10416 | 425 S | PENFSDEDI | 0.993 | unsp |