

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| Chro.20348 | SP | 0.044795 | 0.953241 | 0.001964 | CS pos: 32-33. ISS-DL. Pr: 0.9606 |
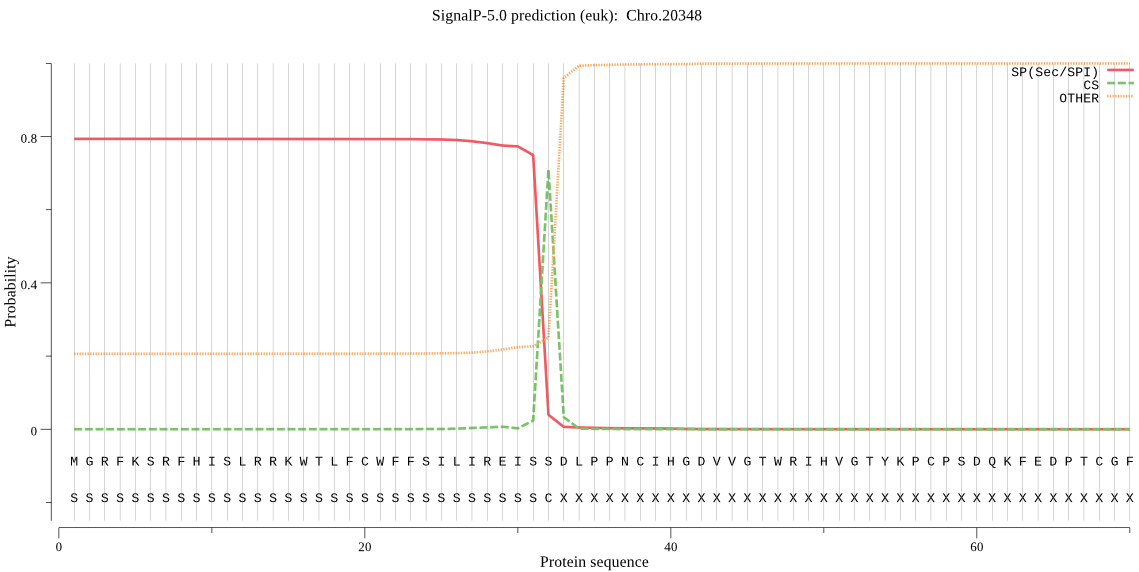
MGRFKSRFHISLRRKWTLFCWFFSILIREISSDLPPNCIHGDVVGTWRIHVGTYKPCPSD QKFEDPTCGFPSPDRDFAHSMLTPTERGLMNNNFKLSFTFDVKFEGSELKVISTQNLDGV DTGKFNDFEKVGTTGNWTVILDQAFTFWTKSYRYTAFFKYINEHEDINASYCYCHTTLLG WWDSYPESVISNQDNKEENKILSLEERSKYHNYPWLVMSEDLKLRRGCWYGMRIFDGDGK MTDRTEWTLKLPRWRSSPLGLPPDHPVNAQNPTIAEYLNEVSAKYSEFDSKNKLWSRSNK FEIGDIQNLDTLPKVRKALKISPHFVQRKKSNDEYITEISSVREEEQISGSKNNTDEPIW MRIRSFDWTNLQHVYGRLGKKVGLVPEVFNQGDCGDCFAVTAATIITSRLWIKYSNEPDI LRKIYASSIQMSECNVYNQGCGGGLITLAFKFAQEVGVRTQDCIDDYAKNLGLKEIYPTP VYTPDEVGIADDGHSFLQTKKGKKFESKKSKIKEIKQQGEDSQEYGQEHKQEHKQGHEQE HDQEHEQEHEQEHEQEHEQEHDQEYEQEQGQEHDQEYQDDYKDMEEHLEGSDSAEHISNV HDYYDDSDDENSSDNEELGDLSGAKHNIPKRFNVIQQLCWDLGGHMGAANTQCRKEIPIT KNIPKSCSKIIKVKEYSYVNNVYGKTTARDIMESLWNEGPVAVSLEPTLEFSLYNSGIFK GFYDPITRQYPWSSIPWHKVDHAMVITGWGWETYGNERIPYWIVQNSWGKRWGEKGFCRI IRGVNELSIEHAAVRASVKIYDSDRKYKAADLQKVHDDSVFQYL
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Chro.20348 | 797 S | AVRASVKIY | 0.993 | unsp | Chro.20348 | 797 S | AVRASVKIY | 0.993 | unsp | Chro.20348 | 797 S | AVRASVKIY | 0.993 | unsp | Chro.20348 | 331 S | QRKKSNDEY | 0.993 | unsp | Chro.20348 | 341 S | TEISSVREE | 0.997 | unsp |