

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| Chro.20366 | OTHER | 0.999965 | 0.000032 | 0.000003 |
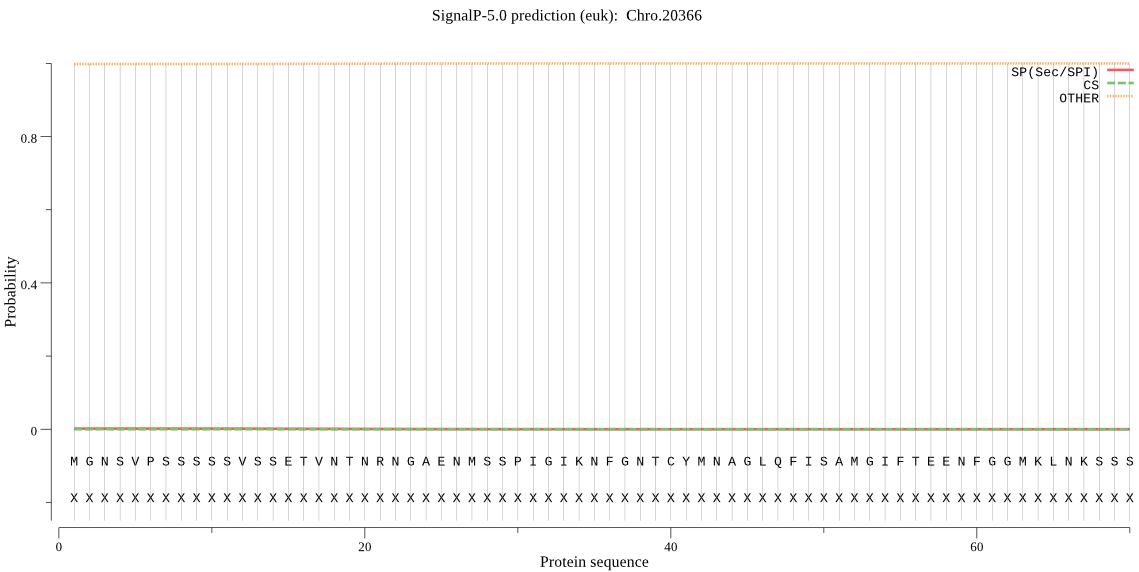
MGNSVPSSSSSVSSETVNTNRNGAENMSSPIGIKNFGNTCYMNAGLQFISAMGIFTEENF GGMKLNKSSSEEVCRSLISILIQLGKPSRITDAISPYQLFKQLRQQNSDLFNQHQQDAHE FIMYILEIIHDGTIKPCNTPKLSNEDLEKLSKKSTRPGNVFWNNHMMRNNSILNNTICGQ FRSRITCENCGNNSDTFDPFWDITLALPEVSDDYEKISIGSCFRKFFEQQNLFHENDEPN YNCSHCKSVVNATRCINISQFPNVLLVTLKRFNNNGEKCNGIVSFKTAGIILKSLTEIGH FKLVAVLQHNGSTLFQGHYISYVFKKEFNGWFKFDDDIVTLVDDILEEDIQAYCLLYVLE SKSALNSNSEQAYSVGIRNQRN
| ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Chro.20366 | S | 4 | 0.608 | 0.019 | Chro.20366 | T | 16 | 0.579 | 0.059 | Chro.20366 | T | 19 | 0.574 | 0.057 | Chro.20366 | S | 7 | 0.507 | 0.077 |
| ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Chro.20366 | S | 4 | 0.608 | 0.019 | Chro.20366 | T | 16 | 0.579 | 0.059 | Chro.20366 | T | 19 | 0.574 | 0.057 | Chro.20366 | S | 7 | 0.507 | 0.077 |
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Chro.20366 | 151 S | LEKLSKKST | 0.996 | unsp | Chro.20366 | 151 S | LEKLSKKST | 0.996 | unsp | Chro.20366 | 151 S | LEKLSKKST | 0.996 | unsp | Chro.20366 | 68 S | KLNKSSSEE | 0.994 | unsp | Chro.20366 | 143 S | TPKLSNEDL | 0.995 | unsp |