

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
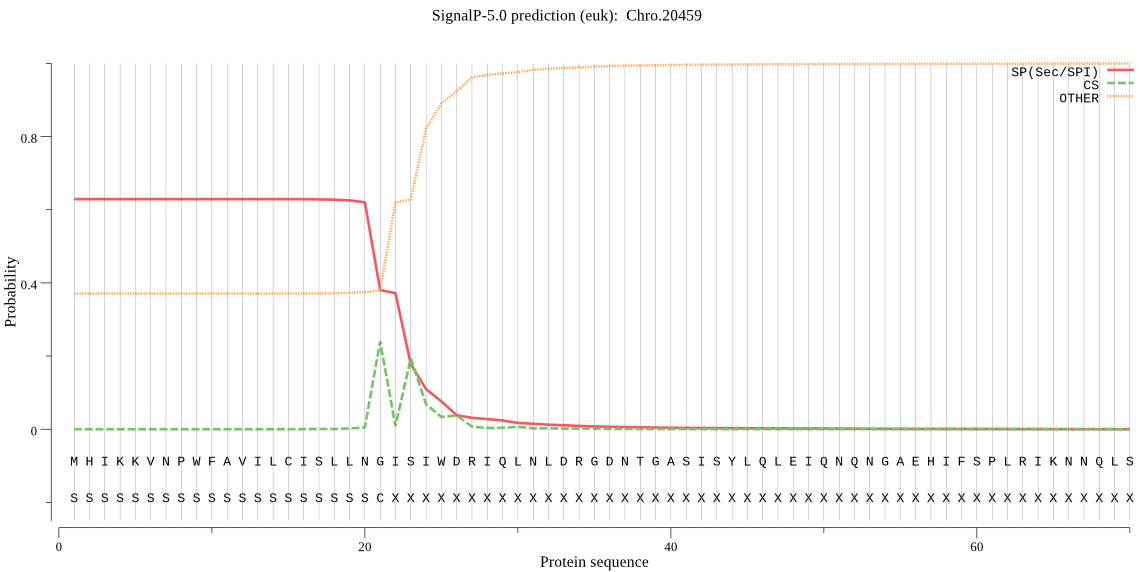
| Chro.20459 | SP | 0.020663 | 0.979253 | 0.000085 | CS pos: 21-22. LNG-IS. Pr: 0.5080 |
MHIKKVNPWFAVILCISLLNGISIWDRIQLNLDRGDNTGASISYLQLEIQNQNGAEHIFS PLRIKNNQLSSLDQQFSSQVKPKAYTTSKGLKTLLVSDNTMLESAFSFGIGCGYYQDPDN LAGLAHLMEHVVFLGSQENPNPVGWDEFLLKKGGAANAYTSADTTIFYVLSPPRELESVM SYFTKMLVNPVIDERSSVSEIDAVNQEHEKNIPNKVRAMIELAMYLAPEECPARKFGTGN KETLYINSKKNNINLKDALKEYHTSCYTSDNASIVIMGPQSNEELVKIADKIDELFTNSG KATKGISKITNTVTSHSDSRTFKNQHSLAPDILSIFESSGNRKSNIGISSTGRITSKQTE SKNKETNQDSIKSKSPNTEIEVVKMPRGSSSPPFVIIYWDSVSDSLSILKENAEWQMLKL IQYFFEDQSQNSIATELQKRKLATALEYFDNTSTQYSVYGLLFTATDDSDQTTSEIAKLT SAYMESLIEQINSKDDKWIQSFYKNYTNMANIKYEYDEERDTAGIASQAAESLLIFPDEP EMALSAFVKPVSMSKDSELSNAQITALKEFVKHLEPSKMKIIKLSDSLNSSSEHNFRFEP YKTEYSISKISLENSENTSKTIETLRSTNKIGELLTCVPGDLSIISFSEDKCPGYKSFEK EIQERKIGNELQPCPILEEEGLSIFWKGPIHTVPTINLTLVQRLPNKDVSNNVRVSLIAN LHAQLQSSKMDYILSSFKLCGLEADISYSRGRFVINVQSYSSNFEDVIEKLSNYLVNGSR LPTKTEFETALTNLKSEILNLSDLMAYDVATDVAQSVYLSNYYSRLQLRESLQKTDITFD EYIEKIKGIFSVGYFDALIVGNIGYEKSIKLVSRMVGSLVNKKIPYSNAIHDGILNVSGD IHIKANNPISSDKNNAVVAHFLTPPVDLIDVSIYSSIGEILNSPFYDTLRTEWQDGYVAF ATTKYETPIISLIGAVQSAEKLSETLVCHMFSALKKVSKDVEEDLKEISKSEFEDKIRWF GLSKYSSQKLDSFTKYIEHFGKLIVSHELCFEKNKLIENATQVFVSEPNIYIEKLNKLIK PSSSRRLVIVELIGNKSPDNKEDELKLVSIANGDIPPTNEECKEILNSEIGKATMMDAIS FKLGSSGSSNGKLRASRNNTSKEENSYVVYDSDIQCNISEENASFASGKDSKEVKTMLNV FGKRMSSMKFSSESKINGNKENSIGNKQNSNSTTKNKRSSCDHSKL
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Chro.20459 | 281 S | MGPQSNEEL | 0.993 | unsp | Chro.20459 | 281 S | MGPQSNEEL | 0.993 | unsp | Chro.20459 | 281 S | MGPQSNEEL | 0.993 | unsp | Chro.20459 | 315 S | NTVTSHSDS | 0.998 | unsp | Chro.20459 | 317 S | VTSHSDSRT | 0.997 | unsp | Chro.20459 | 611 S | ISKISLENS | 0.994 | unsp | Chro.20459 | 1011 S | EISKSEFED | 0.993 | unsp | Chro.20459 | 1156 S | KLRASRNNT | 0.992 | unsp | Chro.20459 | 1160 T | SRNNTSKEE | 0.992 | unsp | Chro.20459 | 1161 S | RNNTSKEEN | 0.996 | unsp | Chro.20459 | 1187 S | ASFASGKDS | 0.995 | unsp | Chro.20459 | 1207 S | KRMSSMKFS | 0.996 | unsp | Chro.20459 | 1239 S | KNKRSSCDH | 0.996 | unsp | Chro.20459 | 1240 S | NKRSSCDHS | 0.996 | unsp | Chro.20459 | 197 S | DERSSVSEI | 0.998 | unsp | Chro.20459 | 199 S | RSSVSEIDA | 0.997 | unsp |