

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| Chro.30255 | OTHER | 0.929292 | 0.059848 | 0.010860 |
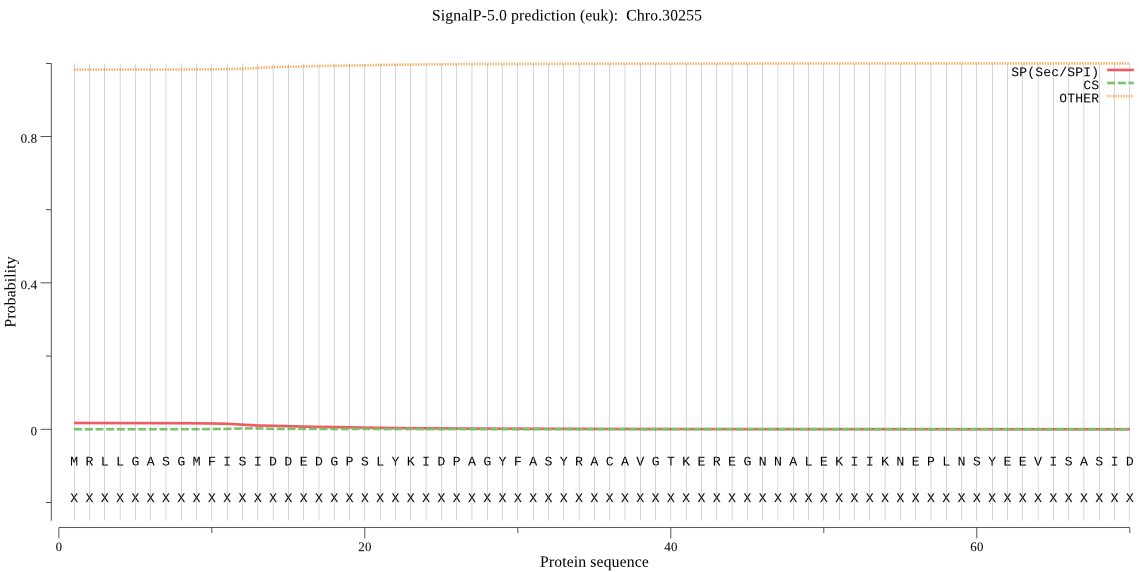
MRLLGASGMFISIDDEDGPSLYKIDPAGYFASYRACAVGTKEREGNNALEKIIKNEPLNS YEEVISASIDCLKTLLGVDFKAEDIEVGVVTKDMPEFRLLSVEEIDNYLTSIAERD
| ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|
| Chro.30255 | 101 S | FRLLSVEEI | 0.997 | unsp |