

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| Chro.30306 | SP | 0.114033 | 0.882795 | 0.003172 | CS pos: 22-23. ACC-AV. Pr: 0.2432 |
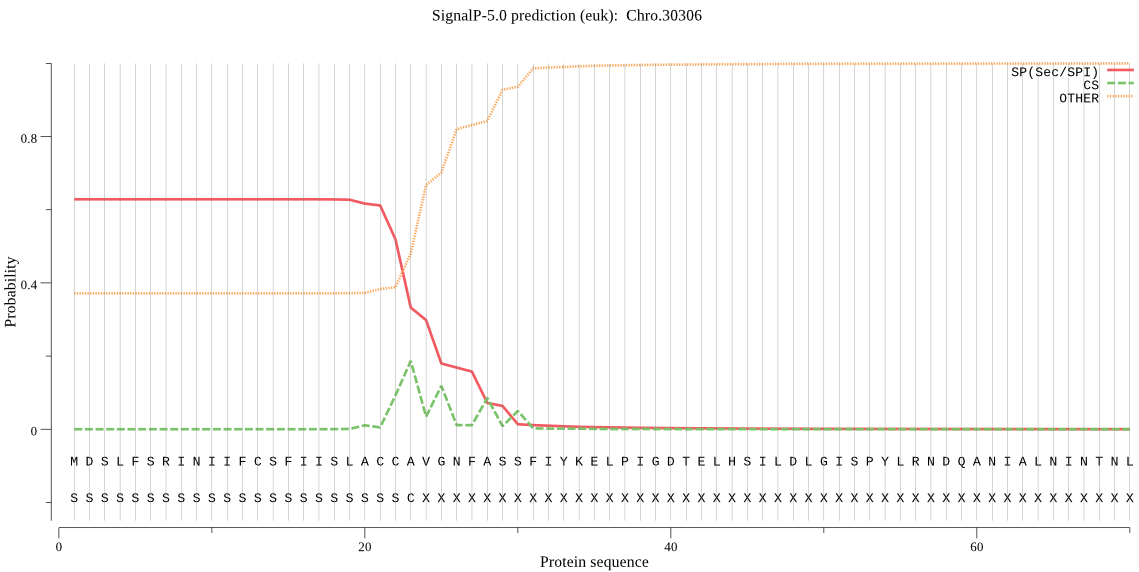
MDSLFSRINIIFCSFIISLACCAVGNFASSFIYKELPIGDTELHSILDLGISPYLRNDQA NIALNINTNLSNSLNWNTNQIFTFIYVSYKNKHQNNYVTVWDDIFSEKKNKTSFSMKGVI NKYPIRDIGRNLRSKSINLNIAFCYMPIVGSIKYHHLKVSIEKKLPVNYFQYK