

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| Chro.30408 | OTHER | 0.999914 | 0.000032 | 0.000054 |
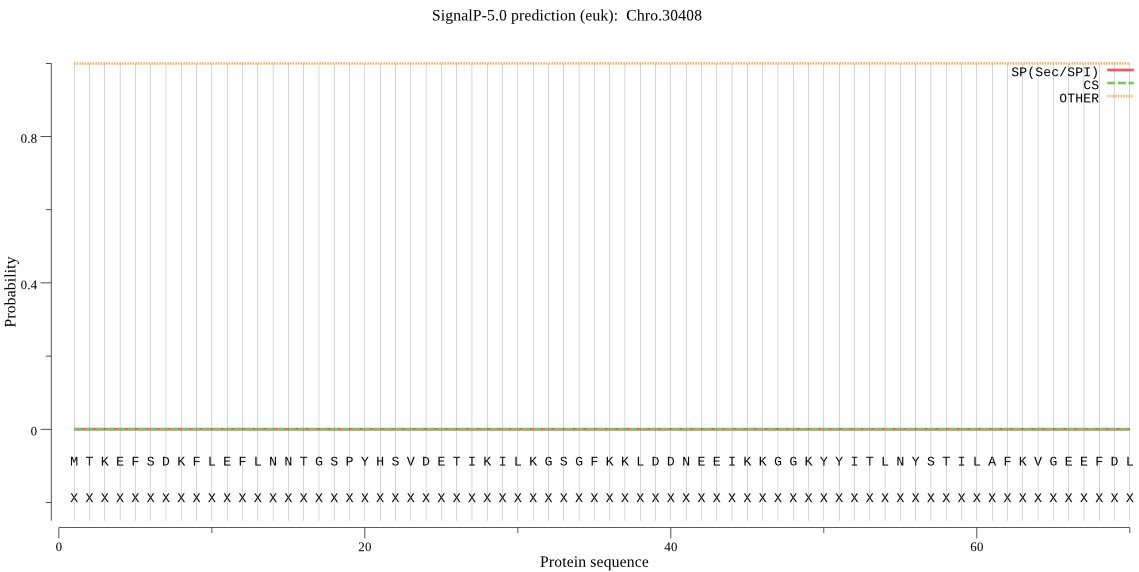
MTKEFSDKFLEFLNNTGSPYHSVDETIKILKGSGFKKLDDNEEIKKGGKYYITLNYSTIL AFKVGEEFDLEDERSMVIITGSHTDSPCLRVRPSSITCNEGYTQLSVSTYGGGIWHTWFD RGLGIAGKVVDNACNEYLIKIKRPICAIPNLAIHLTTSEERSSFVYDKEKHLQPIISKTD TNDKYGEQLLSLPRSLLDEICREINVQPQNISSFDLCLMDSVDSRYVGINDEFIDSPRLD NLGGVFSCFTALIDASESNSSDLLISVAFDHEEVGSVSFSGAHSDFLKQSLKLILAGLEN SSLNNSTPKGMVNANRNSDLEFCKIMKRSIFLSVDMAHSIHPNYPERHQSKHKPSPNNGI VIKTNFNQAYTTDCTTRTFLKTIANNFNIKTQDFLVKNSSPCGSTIGPIVASNLGIRTAD IGACMLAMHSCREFMYYSDIYDLQNFIKVFYMTWSKVKLESKLLE
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Chro.30408 | 95 S | VRPSSITCN | 0.996 | unsp | Chro.30408 | 95 S | VRPSSITCN | 0.996 | unsp | Chro.30408 | 95 S | VRPSSITCN | 0.996 | unsp | Chro.30408 | 158 S | HLTTSEERS | 0.994 | unsp | Chro.30408 | 6 S | TKEFSDKFL | 0.992 | unsp | Chro.30408 | 22 S | SPYHSVDET | 0.996 | unsp |