

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| Chro.30468 | SP | 0.263255 | 0.735367 | 0.001378 | CS pos: 22-23. NFN-YY. Pr: 0.2337 |
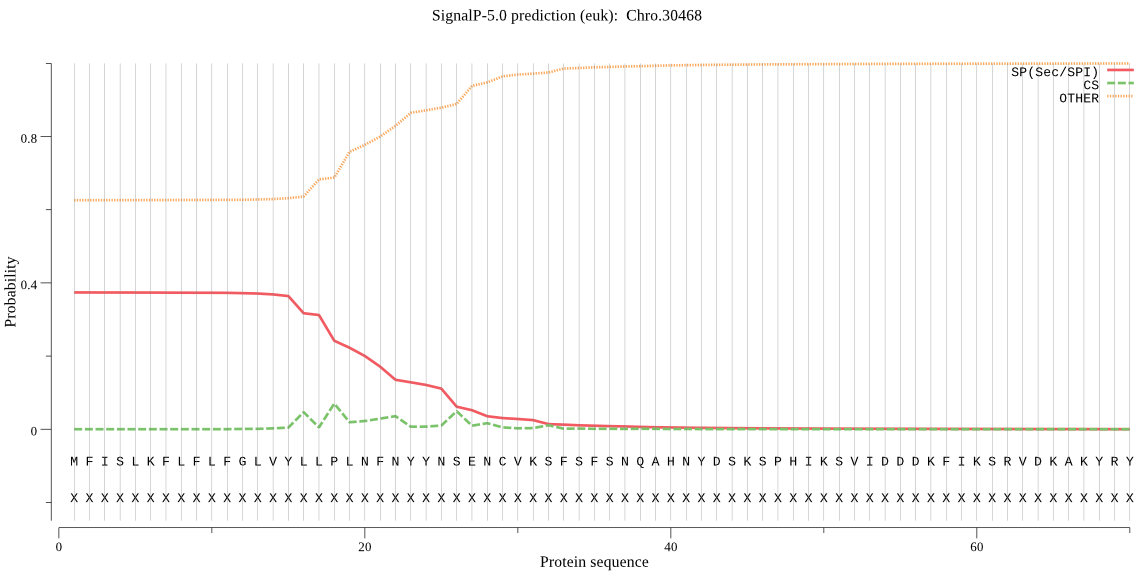
MFISLKFLFLFGLVYLLPLNFNYYNSENCVKSFSFSNQAHNYDSKSPHIKSVIDDDKFIK SRVDKAKYRYIKFKNGLKAFLVSKEDAEKSEVAILVDVGFLYDPPKIIGLSNLVQYSLLL ASYQYPNINEFHNFIKLLNGRIYLDLHEKSTVYSFTIGTEYLSESIFRFSSYFHSPLLNN DTINKAMLTIFSQLNRMKRSEFWARREIEREIIGLNAKFDTFYYGNKNTLLNNPHLSEGE IYEKVRHYFSKFYSPNNMKLAIVGREPLDKLEKYVIQNFAHIRSNGFNIVRIDDNYKYVG NPFIRISKNIVTIRRFKKTGINTINLRFPIEIQVVN
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|
| Chro.30468 | 51 S | PHIKSVIDD | 0.995 | unsp | Chro.30468 | 83 S | AFLVSKEDA | 0.997 | unsp |