

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
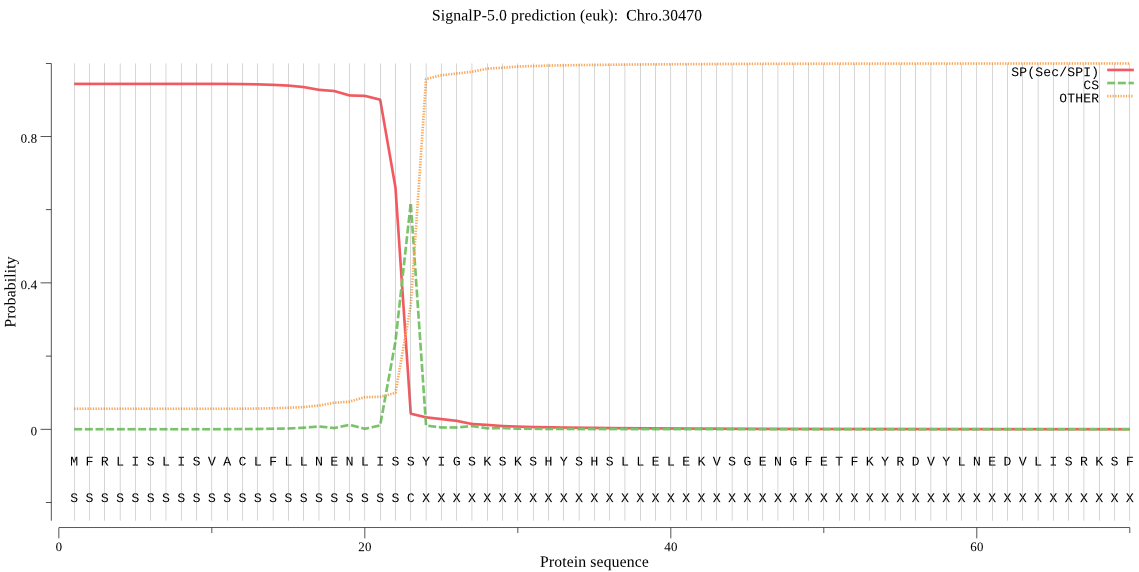
| Chro.30470 | SP | 0.059016 | 0.938893 | 0.002091 | CS pos: 23-24. ISS-YI. Pr: 0.6043 |
MFRLISLISVACLFLLNENLISSYIGSKSKSHYSHSLLELEKVSGENGFETFKYRDVYLN EDVLISRKSFNNGFTLFNIQQPFESSYSDFRITFNSGSSSDPIHLPGIRYFALFIFSKQV NVCMSKVFSEETHANFKIKIDATRSTLIFTFLTEQTNKFFDCFLNTIKNQSYLENSSYFV EILQASEEFEKILDDPSSIEVDKLMIRMIYGNLPSFGCESLQISSIFSESCFKSSIKLNV ELPKYSRQIVNSIFKPHAMSAIFIGKLNNSSLIKGLSSLTKGKLRKTDDLDCNKINKANM AIKNLTSNIILRNGFHTNILRLYIPFETYDLESLASNGQAFILSILNSRHRSGILNFIFM NRFANNVNSYYYSEYTTTILYLEVNLTEKGLQNIPIIIESIVSYLTLIKRTVSSDKLFSE AQNLFNYHLTKSTVILSEAIENYLSSHFIMSKSSSNILNSNINTQFTRANVEYFISKMNL NSITVTLYVPDQLDHLSLFKKNEKSGIFPVETIEPHTNTRVFIFKMADIFPNKLSSLSTK YAVDTFGLQLPNNDLISNISLPFFHSAPIFEKLLHLHKSLDIHYKYLKELGSETIISRTM AYKSITTSGIWFSSNGLFSKSVNLELKFSLRKWSPLLEKFNGEVSSFLILATIHILTAIL NIKLSETLYYTNRLKSNIAFIPAQSFSDMISDPFEIFLVVNAPIDYFTLIMNDVLNTILS FDSYLTEEIVIRAKKAARNTLLRLYEGYTQFEKDLKIVTQIVSSQHCSFLRLGTSLKNDF DIELIRIVYSSLFTSPSIYGLIQGNLTPFHANHILNVFIQGLGHEFFSMEINEENSLNFF NKEVHNNDKNDSNDYAPILEINTSILDISTIPNEYKSMYTHKLDFTADASYSTVVLLIGK LTVPSFIKATAIKEALSNEISSRISSDKKIDFASSLSIVANNFVVVSLSIRSVEFLAGTL NRILDKELNQSISKVLKSHKSKSYVNKILENMNFVDNVFETKFLLSKISILNLVGGTREY LDKMELSDISDLLSDIETVPRIMIATQKVTNLKASKTSLDFIPSGYKDIRSDYKCLLEHP NTVFN
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Chro.30470 | 538 S | LSSLSTKYA | 0.99 | unsp | Chro.30470 | 538 S | LSSLSTKYA | 0.99 | unsp | Chro.30470 | 538 S | LSSLSTKYA | 0.99 | unsp | Chro.30470 | 926 S | SRISSDKKI | 0.996 | unsp | Chro.30470 | 978 S | KVLKSHKSK | 0.992 | unsp | Chro.30470 | 1034 S | SDLLSDIET | 0.995 | unsp | Chro.30470 | 31 S | SKSKSHYSH | 0.994 | unsp | Chro.30470 | 44 S | LEKVSGENG | 0.991 | unsp |