

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
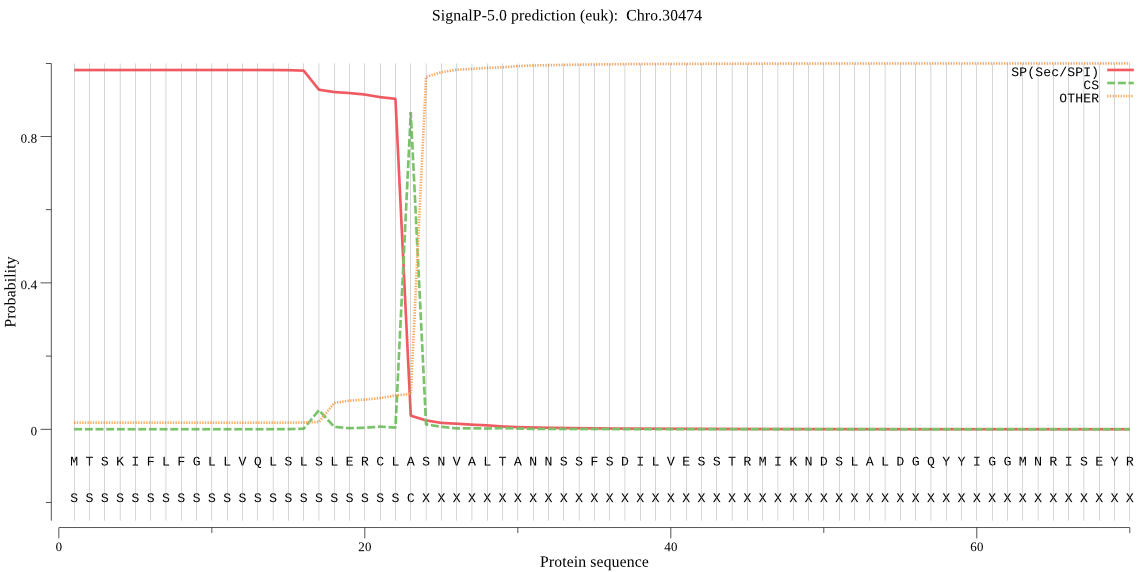
| Chro.30474 | SP | 0.015356 | 0.983284 | 0.001360 | CS pos: 23-24. CLA-SN. Pr: 0.9038 |
MTSKIFLFGLLVQLSLSLERCLASNVALTANNSSFSDILVESSTRMIKNDSLALDGQYYI GGMNRISEYRAKKNINDAYSRFYDEDFIKSVKDSNKYSFIRLKNDIQVFLVSQRSSLFSS ITLGVRVGSSLEPKKFPGLATLLSELLFYDWKRPDVGRETPYDLFISSNSGIFKTKVAPF LTEYHLSIKQEYFSEALIKFCSYLKSFSPKRIHLDPAMEALQSDFEALMGMSFIKLKQIL KELSVEGHVNHGFHMGNMKRLMANIDFDTEVLLFELIKFYGSYYSSNLMTISIVSDKSID ELEGLARTFFDEIPNQSKQLITPFDLSNEIIHPYIDLNNKVIQVKSAEENTYFTVIFPIP HQSPLWKYKPAEYISFFFTDYSNKSLYSYLRGIGIILGLKTMIEVNDNGFSNFIIRFKLN SKGEKAIVKILEITLSFLKLIKEVSISETIINQIRKKRQSILEVSIDALYSDLSRKIVNT FLRTNCSPPEVLHAGVSMNKIEFNHVKEIIDHMKYDNLILILEKKEFKKSASNILLSSQF FLDSKWKSARKSISSFFRSVNTAFHRTKVGTFPKFLKEEYLGGEYLLEDIPSKVIEVLQS VNSTLANDILKLQMPEIDPKFPRNFVIYTEDVPKKEFPVSLYYAVKEFKQAKNLTEISLS EASNNSTDFSYLNLSAPINPRDLVVALSTQKMDSISRSFCYLPTSFNPDVMISVRLQIPN NPPKEFSCPEKLAILMFLFQNTLYDVLPSHLNENFSFERSYHHWELQEFFYGIEFSWRGY SSIFPDSLNEIENILSIFQTSIKQSSFQNAKQRFQDIINSFKSENDNLRLLSLSYQLLDP GFMKISRLEDEMSKITIQSLIDFANFFIKNFSISGSVLGNATPIQIKYYLSSFVNTVRGM DSSSLELSLDTTENMFNSTKQVNSDINEDFNTLGFEEFSFEEKGRMLRSGSKKSTSKDLK NDNNNQIGKPNEVTHTNTYNAMNIENLGYTRNSFSDMSRLPYNYSNSYFFYINSSKNSKD NMVLLQIHYGFYSEQNLAFLQIVSELNSYKYFIEFSDKVCPDCSLKVLPRIILGKYLVLE FKLQSPTKNIRELGELLNNFFLTYYSRPSKIISKSEFNKAKEFLLLLSRNARLIENNLGG QITSKTNKIYYEGSESEDLFTLASLHMGASKHTHFSCSWKKDYLEFIDALTFDQFYKYWQ YFGSSSKLFISYQSQNTNYELFESLESYLPQGFTKLLFAESLYDINEQQDIFNT
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Chro.30474 | 160 T | VGRETPYDL | 0.993 | unsp | Chro.30474 | 160 T | VGRETPYDL | 0.993 | unsp | Chro.30474 | 160 T | VGRETPYDL | 0.993 | unsp | Chro.30474 | 208 S | LKSFSPKRI | 0.996 | unsp | Chro.30474 | 244 S | LKELSVEGH | 0.993 | unsp | Chro.30474 | 295 S | ISIVSDKSI | 0.992 | unsp | Chro.30474 | 298 S | VSDKSIDEL | 0.996 | unsp | Chro.30474 | 460 S | KKRQSILEV | 0.995 | unsp | Chro.30474 | 552 S | SARKSISSF | 0.995 | unsp | Chro.30474 | 694 S | QKMDSISRS | 0.996 | unsp | Chro.30474 | 823 S | NSFKSENDN | 0.994 | unsp | Chro.30474 | 951 S | LRSGSKKST | 0.997 | unsp | Chro.30474 | 993 S | YTRNSFSDM | 0.997 | unsp | Chro.30474 | 67 S | MNRISEYRA | 0.994 | unsp | Chro.30474 | 90 S | DFIKSVKDS | 0.995 | unsp |