

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
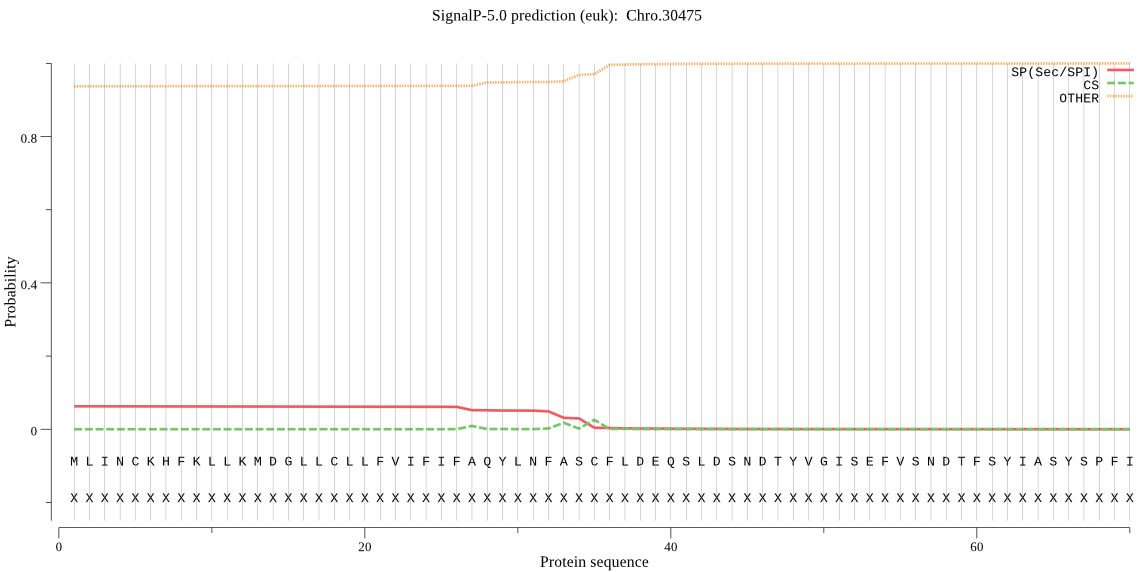
| Chro.30475 | SP | 0.475388 | 0.523758 | 0.000854 | CS pos: 35-36. ASC-FL. Pr: 0.7431 |
MLINCKHFKLLKMDGLLCLLFVIFIFAQYLNFASCFLDEQSLDSNDTYVGISEFVSNDTF SYIASYSPFILDGFSDMNSFSKMRSIYYNVNDFEEVDNKDFVRMANENGEYKFITLENKI KVFLVSKNILYKSSIAISIGYGDFNEPSHMKGLSVLLKNIVVSKIKFKTNKRTNPLYFKN LENTVEGRVHGHGTEINIEVFNEDFVDVLHTLSEVLASPLELDFHTLSSSRKITREEIRA RCISMENLRGAFVENDFCKKSGYKYLYAFTDSNVNSKDQFKRGYRIGANSKKVDEFSDEY SSLDSILNEHFNKYYCSNLMTLAIVSNESMHYLLTLVKTFFGKIRSSNFNTNDLEKIYNF NENPYLKHVGNVVAVKQSSVNLELKLIFPIPYQKNLWRYKLTEYISFFLDDESEEGLTGV LRSRGWITSLSSGCETNESGYSNFVVLITLQKEGVRHIMQILEALLSTIKLISKQDFSEE TLFHIREESSIELKNLYLDLDTDQASFILRSYLGTGCFPKDVLVAPFKMDQVEGRHIKKL LNFIRADNMVISHPVKSLNNGASYYLRSSESFNNSFLFNFEKMVFKSFYNLSLFFLKWFK TGPEQGDCRFYKKLHYSEEKLPQYLQRRLSSIDERVALEELKIQLFDPSLESNSQIYIND QLKTINYPITLRKGIEDKLLNDNFFRNSKTEANSKTSQHIDDKELYRNDFKLNVDHNLID LKEGNSSHESYNNKLSIDYADEYEGFMEENKIGELDYETGKKIDSLYPEFDMDSIFYMPL HNKLPTLSVTFDITIPFDVNNSTDLLLLNMNRQKLLLLSFLFKYSMNLSLADSNSDGVIK VENYTDFMNIDVLPGIQLSWDGITKYFDDFFMVFAHNLVNYRTIITNCYFQKSITALKKL LERLSYASNEDYSLMLLMQITHLESVKSWSLENDAKHLTLEDVKLFGKILLKYGEVHGVA IGNATPSQIYARINKFIKMIRPSASLLNSFKQIHSGQFSMTFDTKSYGWISDKDLTDEMI KRSYMGPMLMNAENKGNLFNSKFINMELLPSMYHKNYFFKQRASSSDPFNTVLLYIYLGN TERNMVLLDLLELVDFHKVLNSFIANKKSLRYSSIQIGSLFITRDVASFQIKVTFLSNKI ISMVNVILEFFDIYFLNPEKVFSKRDFLSLKSMLLSNFQKVSLKPSKLVQMHYERIIRGN LGNDWQIKETVLLRNFCYLGFLELWSSFRTAPTILVIIQSNRNQDDQSNQLSEFVPEGYT RLKSIYQFKDSFNVH
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Chro.30475 | 290 S | IGANSKKVD | 0.993 | unsp | Chro.30475 | 290 S | IGANSKKVD | 0.993 | unsp | Chro.30475 | 290 S | IGANSKKVD | 0.993 | unsp | Chro.30475 | 630 S | QRRLSSIDE | 0.998 | unsp | Chro.30475 | 833 S | SLADSNSDG | 0.992 | unsp | Chro.30475 | 908 S | LSYASNEDY | 0.996 | unsp | Chro.30475 | 1011 S | YGWISDKDL | 0.994 | unsp | Chro.30475 | 1064 S | KQRASSSDP | 0.998 | unsp | Chro.30475 | 1163 S | EKVFSKRDF | 0.996 | unsp | Chro.30475 | 218 S | EVLASPLEL | 0.99 | unsp | Chro.30475 | 244 S | ARCISMENL | 0.994 | unsp |