

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| Chro.40078 | OTHER | 0.767812 | 0.155408 | 0.076779 |
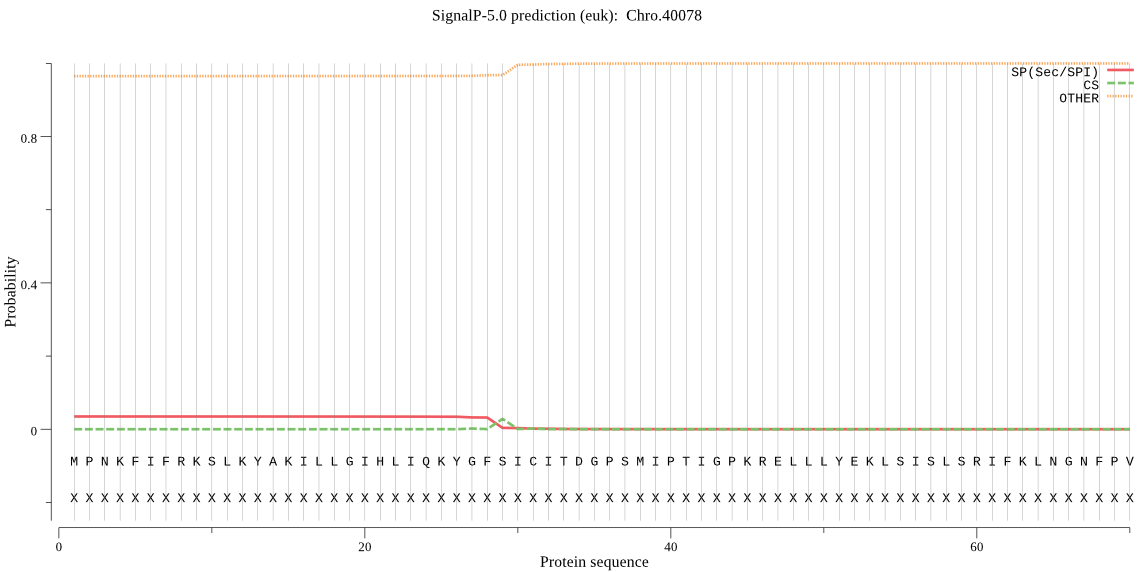
MPNKFIFRKSLKYAKILLGIHLIQKYGFSICITDGPSMIPTIGPKRELLLYEKLSISLSR IFKLNGNFPVNRNDIIIANSVENPEILVCKRSIAKVXYD
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|
| Chro.40078 | 10 S | IFRKSLKYA | 0.995 | unsp | Chro.40078 | 80 S | IIANSVENP | 0.991 | unsp |