

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| Chro.50094 | OTHER | 0.995165 | 0.002505 | 0.002330 |
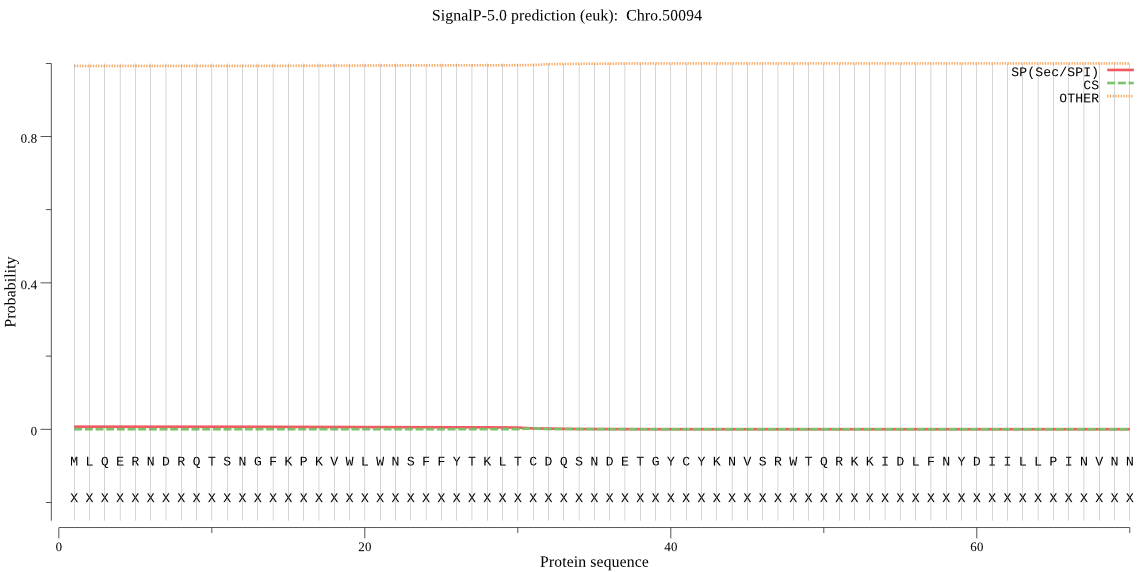
MLQERNDRQTSNGFKPKVWLWNSFFYTKLTCDQSNDETGYCYKNVSRWTQRKKIDLFNYD IILLPINVNNVHWTLGVVNFKLGYIQYIDSLGGQFQDHLGCTKMSSIFFQNMNRYIQDEY FDKKKEKFPGQLKHFTRFSEPVPQQNNGSDCGVFTCMFAECISEGRSFDFDTTQIDRIRE VMLVECIKNEIF