

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| Chro.50185 | OTHER | 0.999179 | 0.000656 | 0.000165 |
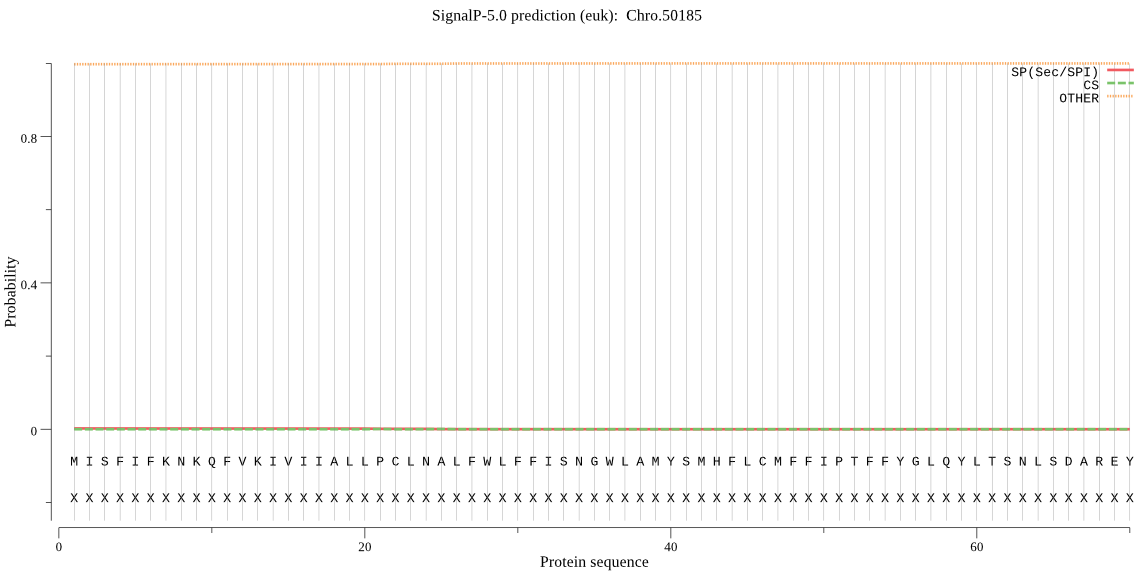
MISFIFKNKQFVKIVIIALLPCLNALFWLFFISNGWLAMYSMHFLCMFFIPTFFYGLQYL TSNLSDAREYGKNFDILKCICVCIFTSVFGTTCLFLVSELSKIIPTFDLLNIEEIKNGLL KEGIIKDPTNKIGFSTWFSSIYFSIVNPIIEELFWRSFVYKELIISLGVRNETNPQSRFN INEIGANYSDFLIARDHSSNLFESYDVESGQIGYKANTELASIICSVLYSLYHFFVLIRF SSIHCSILSTISLAIIGRALLYVNRKFHTIYSIYIHIGMDISIVIFLILSLK