

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| Chro.50200 | OTHER | 0.993357 | 0.006322 | 0.000320 |
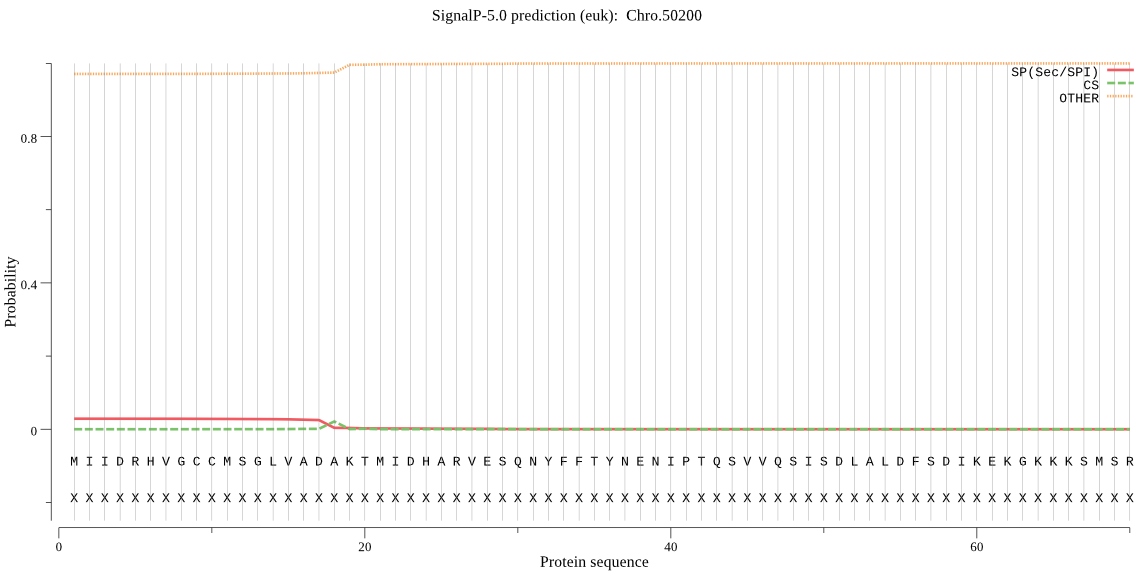
MIIDRHVGCCMSGLVADAKTMIDHARVESQNYFFTYNENIPTQSVVQSISDLALDFSDIK EKGKKKSMSRPFGVAMLIAGADSDGSSSLWMTDPSGTYTQYSAAAIGTAQEGAETILLEN YNSNMSLKEAEDLALIVLRQVMEEKINSVNVEVAAVKEKKFIIYDNKEIQRVLDRLPPPT HIVPSQLS
| ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Chro.50200 | S | 188 | 0.576 | 0.029 | Chro.50200 | S | 185 | 0.523 | 0.237 | Chro.50200 | T | 92 | 0.504 | 0.110 | Chro.50200 | T | 99 | 0.500 | 0.045 |
| ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Chro.50200 | S | 188 | 0.576 | 0.029 | Chro.50200 | S | 185 | 0.523 | 0.237 | Chro.50200 | T | 92 | 0.504 | 0.110 | Chro.50200 | T | 99 | 0.500 | 0.045 |
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|
| Chro.50200 | 67 S | GKKKSMSRP | 0.992 | unsp | Chro.50200 | 126 S | NSNMSLKEA | 0.997 | unsp |