

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| Chro.50347 | OTHER | 0.996239 | 0.003317 | 0.000443 |
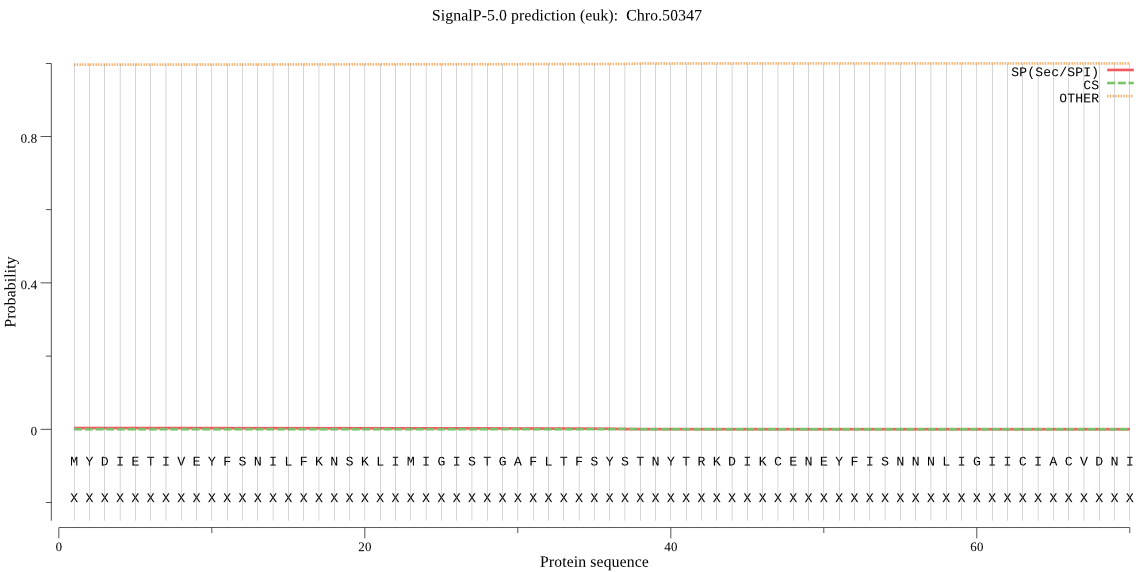
MYDIETIVEYFSNILFKNSKLIMIGISTGAFLTFSYSTNYTRKDIKCENEYFISNNNLIG IICIACVDNIPESYILDFSNEQLNEFNELGYTKINTKIPIVKINHINKVKDDLKLISRNY LESYNIFPTYNFLLENKEKVVNYPILLIHGTDDQSVPFQMSLNLLNLNNNFSNKNNRYLR LLKINNGNHLLSNSKHIKKAQNEISNFVFEIL