

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| Chro.60410 | SP | 0.231052 | 0.664983 | 0.103965 | CS pos: 25-26. LIG-VN. Pr: 0.6069 |
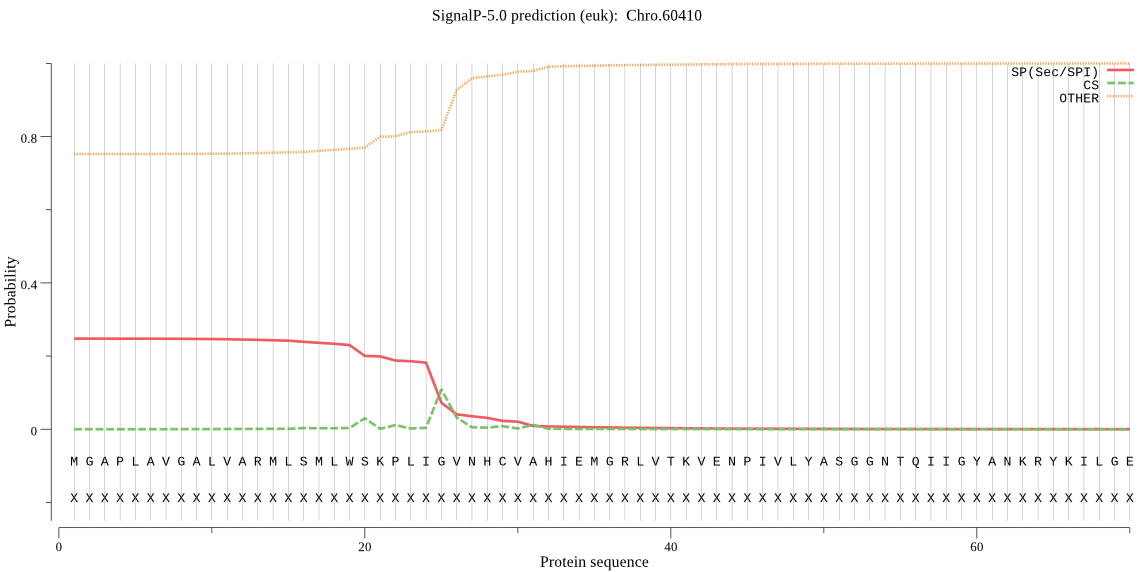
MGAPLAVGALVARMLSMLWSKPLIGVNHCVAHIEMGRLVTKVENPIVLYASGGNTQIIGY ANKRYKILGETLDIAIGNCIDRFARVMKLDNYPAAGYHIEQMAKKGKNLISLPYVVKGMD LSFSGILTFGEELIAEKQKEFNNDKQKLHSFYQDFCFSLQETLFAMLIEVTERAISLLNS DSILLVGGVGCN
| ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|
| Chro.60410 | 176 S | ERAISLLNS | 0.993 | unsp |