

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| Chro.60563 | OTHER | 0.999939 | 0.000054 | 0.000007 |
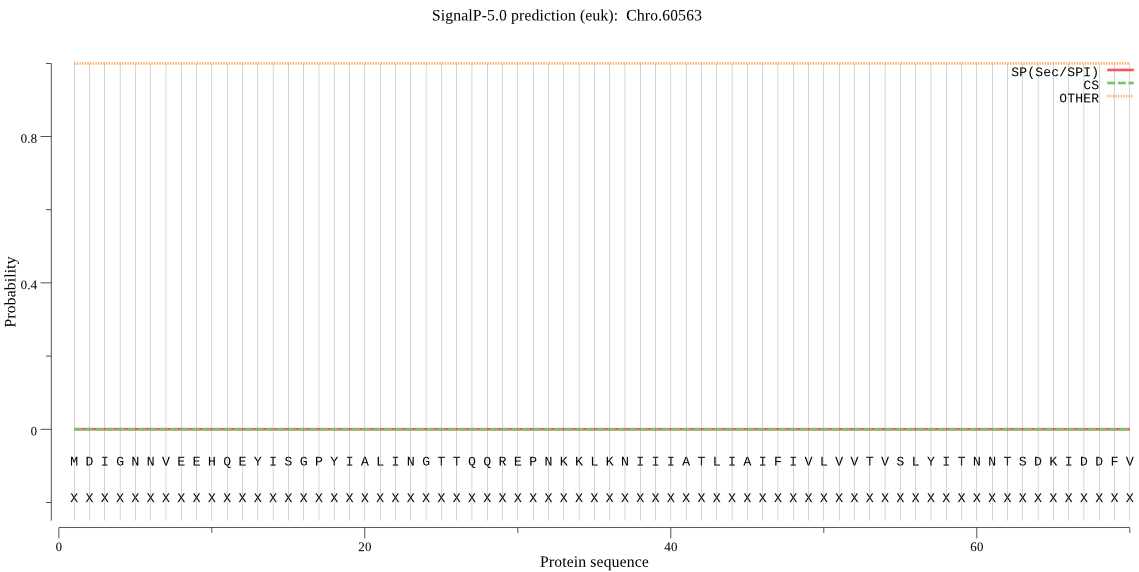
MDIGNNVEEHQEYISGPYIALINGTTQQREPNKKLKNIIIATLIAIFIVLVVTVSLYITN NTSDKIDDFVPGDYVDPATREYRKSFEEFKKKYNKTYSSMEEENQRFEIYKQNMNFIKTT NSQGFSYVLEMNEFGDLSKEEFMARFTGYIKDSKDDERVFKSSRVSASELEEEFVPPNSI NWVEAGCVNPIRNQKNCGSCWAFSAVAALEGATCAQTNRGLPSLSEQQFVDCSKQNGNFG CDGGTMGLAFQYAIKNKYLCTNDDYPYFAEEKTCMDSFCENYIEIPVKAYKYVFPRNINT LKTALAKYGPISVAIQADQTPFQFYKSGVFDAPCGTKVNHGVVLVGYDMDEDTNKEYWLV RNSWGEAWGEKGYIKLALHSGKKGTCGILVEPVYPVINQSI
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Chro.60563 | 166 S | SSRVSASEL | 0.998 | unsp | Chro.60563 | 166 S | SSRVSASEL | 0.998 | unsp | Chro.60563 | 166 S | SSRVSASEL | 0.998 | unsp | Chro.60563 | 85 S | EYRKSFEEF | 0.997 | unsp | Chro.60563 | 153 S | YIKDSKDDE | 0.996 | unsp |