

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
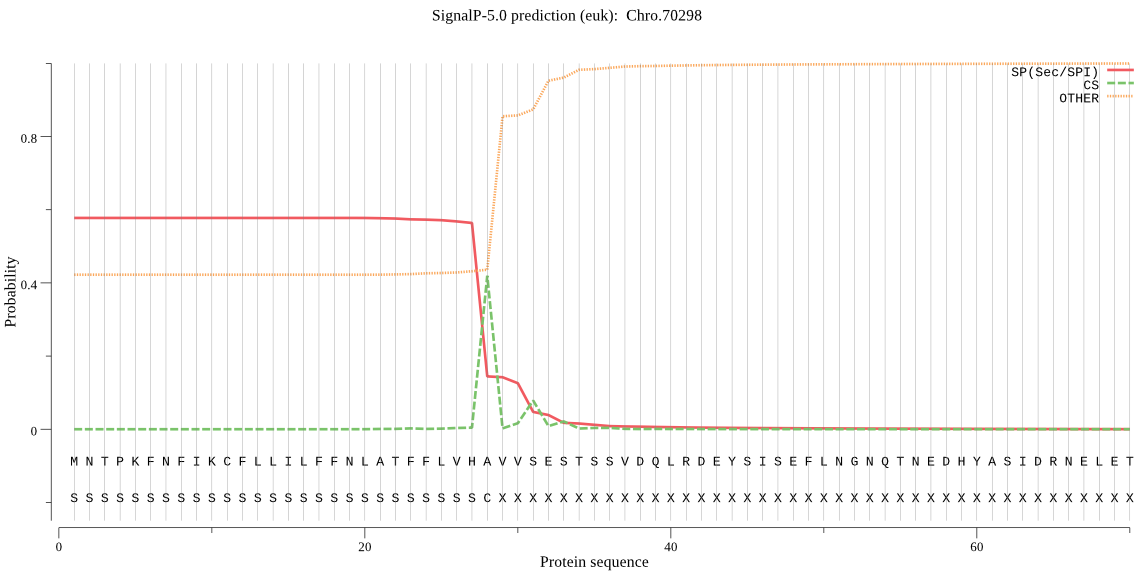
| Chro.70298 | SP | 0.187192 | 0.812588 | 0.000220 | CS pos: 28-29. VHA-VV. Pr: 0.7161 |
MNTPKFNFIKCFLLILFFNLATFFLVHAVVSESTSSVDQLRDEYSISEFLNGNQTNEDHY ASIDRNELETRPSFDSNRESLNAHLENKLTGVNKQSFVEFQKGKLSKNDVDIEELIFEIV KLALDGNGSRVLPSLELIDFLQKIIETSKYNTVTTLDYILAFISKEKFDTSITHDINRLR NFTEYLSYLLKYNDNGPNRMLNSGKETLSLIKTRIHSSIWGMEELIRTNKIMDYEIQNLG GKTTYSSQPVIGEITNEINSNQPRAVFSYKLVKQTIIEMLGTNINKVYSKLKRIYGKITY SGSSKSNKSISLNLLYAIYLSAIMINDLYNQGLSSNLLSPGFIFMVLLRDDIEHLPNNWE ERLIKRSISYGINEIDIKIQDSKYSNLESIITKGFTHSECNASVLRSNIQVVFRKYGIEY FKFMNEALKKYKTFFVPSTDGRINSISWFNNYSVNGYTNKLNKKWFVDINELVIENGVTK AVDFMDEFHLLEISLSRSGLSSAIIVSESKLLKGTLVEYLAYRIINGNSSIDLRGYRIIS IHLESLLESCKNTKKSLTEQIKIKFDELMGAYDGKIIVFTDNLFSSFETSTGSKRLYDIM KHYIVLGTLKVIATLSNENYKILTEKEIEVKSIFYTIEMKELNGIVSEVFISGLRYQLEL STGIFINNDVIRVSVLMCHKYIENCVLPDDAVELINFAISMAKNEQFLEIPSAIHGIEDF ISHSRIGAQVSKMRDYESNSIISSNRSRLSEVLAMHLRMRNRIVSLALKIRPYLTNFRYL KTQLYFMMLINLNYQTSCVNKPVEDEFKLLENREKIIPNFSEELSVTQLTSLYRKNLLKY KQINNKSYNESINDEVSAITESQLDPMFFDDIHQVVKDKQKEVADMKSLILEMAFNYNSL YSPLGMGEIDASHIAFIISEKYGKSITKLLDEIEYRKLPERMKDRLSEILSKYVIGQKSA IDYVSFHLGVELLRNNKNNPRCLLFVGPSGSGKKTFALALQTTLAESSVLFYDFSYDITM YSHFKILKSSDFADADAAKRLLGDNPKTGIISEELKQTGKTVFFFEHIENMHPDVIKLIL DILHNKDNINHKIGNLGSSTFILTTKVGSEIILKNPDQIDRIKIRVDIIKKLRETFSSDL VRLIQNVVLFRPFTKEEALKILELNFSEFSVLIKKNYSTIFIQPSLLVLNKIVDKKYSPE LGYKSVLDFFEKEVKEKVLQLIKKGILVPYTTFKITIRDISSVEDPNGFKIDFAIVKKSI NSR
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Chro.70298 | 76 S | PSFDSNRES | 0.991 | unsp | Chro.70298 | 76 S | PSFDSNRES | 0.991 | unsp | Chro.70298 | 76 S | PSFDSNRES | 0.991 | unsp | Chro.70298 | 369 S | KRSISYGIN | 0.992 | unsp | Chro.70298 | 556 S | NTKKSLTEQ | 0.995 | unsp | Chro.70298 | 947 S | KDRLSEILS | 0.994 | unsp | Chro.70298 | 1241 S | IRDISSVED | 0.996 | unsp | Chro.70298 | 1242 S | RDISSVEDP | 0.996 | unsp | Chro.70298 | 35 S | SESTSSVDQ | 0.993 | unsp | Chro.70298 | 60 Y | NEDHYASID | 0.991 | unsp |