

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
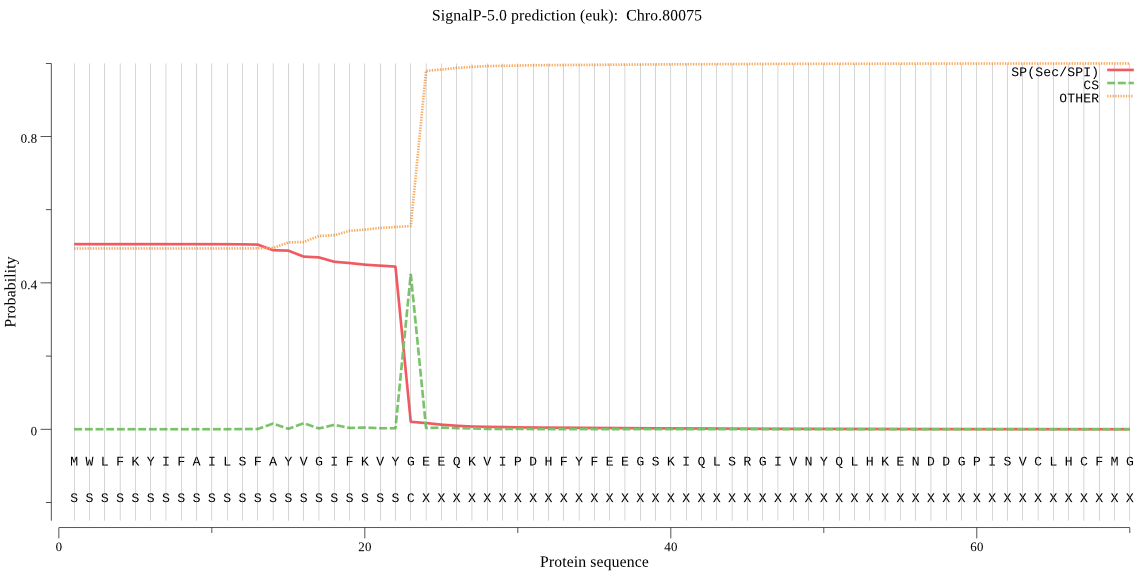
| Chro.80075 | SP | 0.011184 | 0.988698 | 0.000118 | CS pos: 23-24. VYG-EE. Pr: 0.9075 |
MWLFKYIFAILSFAYVGIFKVYGEEQKVIPDHFYFEEGSKIQLSRGIVNYQLHKENDDGP ISVCLHCFMGTISDCSSISKNLAKNGYRTLRFDFYGHGLSQYKNFGQYSVDDYVDQTMEL LEKLGLYNITAISEEELHSSSFTPKLHVIGTSLGGFVAMRIAQRFPKHIGKLVLDAPPGL LKKKVAPYLQYSIINYPLQLFANIYSPVWGCYQFMDPPSENLSSPKLDFRAKHYCKQILL TSLQLFLGINLWNNQQIYQEFSKVDVPTLFFWGAEDRLCPLSSAITILNEYLPNTKIIVY ENCKHRCSKYCKEQFVEDVIKYFKNELDQELVSMSQYYNSVHDMVHSSEKRSLPSIRGTK TDTLLEPNSISEDTCSTSTSGSQEIAVQLASSTEDNTETGSSMSGSGNFNDDFCKTKN
| ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | |
|---|---|---|---|---|---|---|---|---|---|---|
| Chro.80075 | T | 393 | 0.518 | 0.062 | Chro.80075 | T | 379 | 0.501 | 0.077 |
| ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | |
|---|---|---|---|---|---|---|---|---|---|---|
| Chro.80075 | T | 393 | 0.518 | 0.062 | Chro.80075 | T | 379 | 0.501 | 0.077 |
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|
| Chro.80075 | 355 S | RSLPSIRGT | 0.993 | unsp | Chro.80075 | 392 S | QLASSTEDN | 0.997 | unsp |